5I4A
 
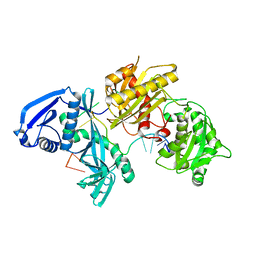 | | X-ray crystal structure of Marinitoga piezophila Argonaute in complex with 5' OH guide RNA | | Descriptor: | Argonaute protein, RNA (5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*U)-3') | | Authors: | Doxzen, K.W, Kaya, E, Knoll, K.R, Wilson, R.C, Strutt, S.C, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | A bacterial Argonaute with noncanonical guide RNA specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4KM5
 
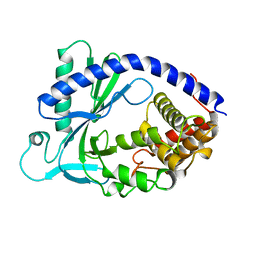 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
7S38
 
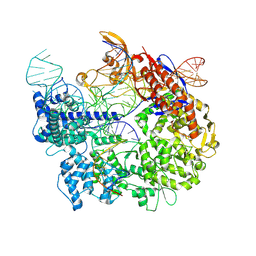 | | Cas9:sgRNA:DNA (S. pyogenes) forming a 3-base-pair R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S37
 
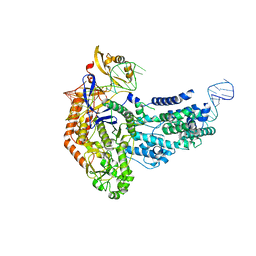 | | Cas9:sgRNA (S. pyogenes) in the open-protein conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Single-guide RNA | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S3H
 
 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-protein/linear-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-06 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S36
 
 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, closed-protein/bent-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7THB
 
 | | Crystal structure of an RNA-5'/DNA-3' strand exchange junction | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*TP*AP*AP*GP*CP*AP*GP*CP*AP*TP*C)-3'), RNA (5'-R(*AP*GP*CP*UP*UP*AP*C)-3') | | Authors: | Cofsky, J.C, Knott, G.J, Gee, C.L, Doudna, J.A. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of an RNA/DNA strand exchange junction.
Plos One, 17, 2022
|
|
4ZT0
 
 | |
4ZT9
 
 | |
2X04
 
 | | Crystal structure of the PABC-TNRC6C complex | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1, SULFATE ION, TRINUCLEOTIDE REPEAT-CONTAINING GENE 6C PROTEIN | | Authors: | Jinek, M, Fabian, M.R, Coyle, S.M, Sonenberg, N, Doudna, J.A. | | Deposit date: | 2009-12-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Insights Into the Human Gw182-Pabc Interaction in Microrna-Mediated Deadenylation
Nat.Struct.Mol.Biol., 17, 2010
|
|
5K4B
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 1 | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4D
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4C
 
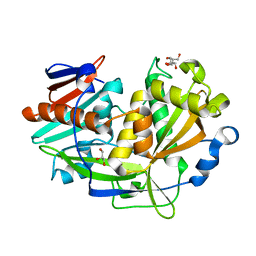 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 2 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D, GLYCEROL | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
6MCB
 
 | |
6NY3
 
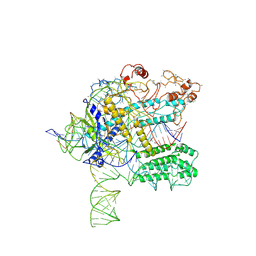 | | CasX ternary complex with 30bp target DNA | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6NY2
 
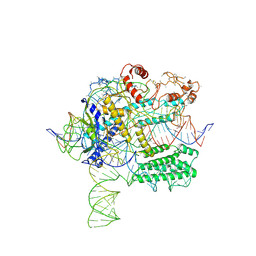 | | CasX-gRNA-DNA(45bp) state I | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6NY1
 
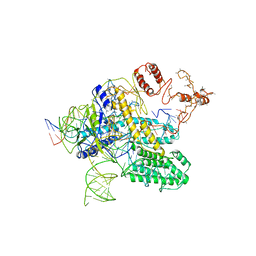 | | CasX-gRNA-DNA(30bp) State II | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
7KFU
 
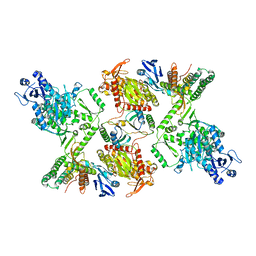 | | Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
7KFT
 
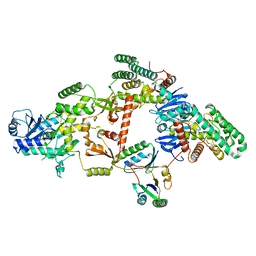 | | Partial Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
1DRZ
 
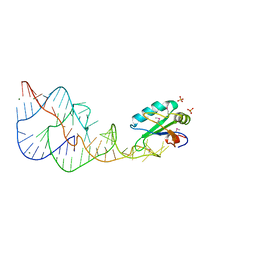 | | U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX | | Descriptor: | MAGNESIUM ION, PROTEIN (U1 SMALL RIBONUCLEOPROTEIN A), RNA (HEPATITIS DELTA VIRUS GENOMIC RIBOZYME), ... | | Authors: | Ferre-D'Amare, A.R, Zhou, K, Doudna, J.A. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a hepatitis delta virus ribozyme.
Nature, 395, 1998
|
|
1DUL
 
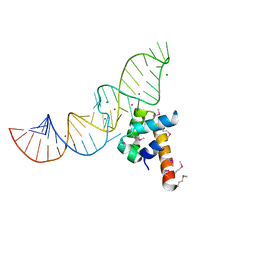 | | STRUCTURE OF THE RIBONUCLEOPROTEIN CORE OF THE E. COLI SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5 S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Rambo, R.P, Lucast, L, Rha, B, Doudna, J.A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-28 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ribonucleoprotein core of the signal recognition particle.
Science, 287, 2000
|
|
1KXK
 
 | |
1KH6
 
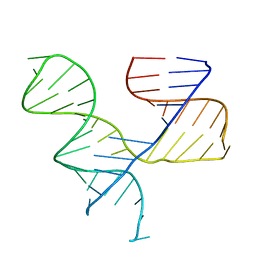 | | Crystal Structure of an RNA Tertiary Domain Essential to HCV IRES-mediated Translation Initiation. | | Descriptor: | JIIIabc | | Authors: | Kieft, J.S, Zhou, K, Grech, A, Jubin, R, Doudna, J.A. | | Deposit date: | 2001-11-29 | | Release date: | 2002-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation.
Nat.Struct.Biol., 9, 2002
|
|
1L8V
 
 | |
2Y8W
 
 | |
