3KX7
 
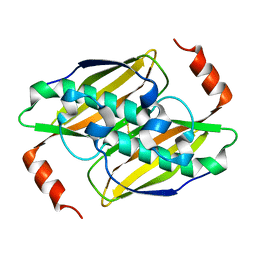 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - apo wild type FlK | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3N86
 
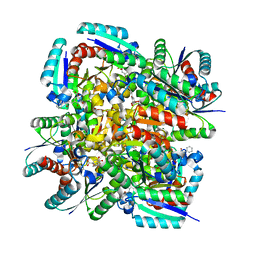 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 4 | | Descriptor: | (1R,5R)-1,5-dihydroxy-4-oxo-3-[3-oxo-3-(phenylamino)propyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N76
 
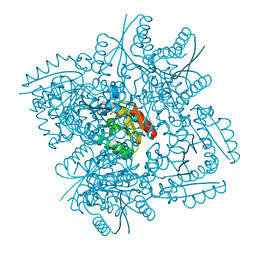 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with compound 5 | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N87
 
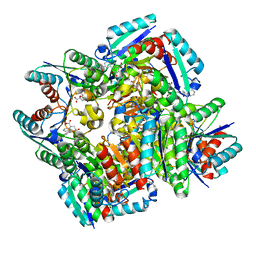 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 3 | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[3-(phenylcarbonyl)phenyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3KUV
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42S mutant in complex with acetate. | | Descriptor: | ACETATE ION, Fluoroacetyl coenzyme A thioesterase | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3N8N
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 6 | | Descriptor: | (1R,4R,5R)-3-(tert-butylcarbamoyl)-1,4,5-trihydroxycyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N7A
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 2 | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
8G6P
 
 | | Crystal structure of Mycobacterium thermoresistibile MurE in complex with ADP and 2,6-Diaminopimelic acid | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Rossini, N.O, Silva, C.S, Dias, M.V.B. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of Mycobacterium thermoresistibile MurE ligase reveals the binding mode of the substrate m-diaminopimelate.
J.Struct.Biol., 215, 2023
|
|
9AUE
 
 | | Crystal structure of the holo form of GenB2 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, C-6' aminotransferase, ... | | Authors: | Oliveira, G.S, Bury, P.S, Huang, F, Li, Y, Araujo, N.C, Zhou, J, Sun, Y, Leeper, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
9AU3
 
 | | Crystal structure of GenB2 in complex with G418 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, C-6' aminotransferase, ... | | Authors: | De Oliveira, G.S, Bury, P.S, Huang, F, Li, Y, Araujo, N.C, Zhou, J, Sun, Y, Leeper, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
9B0C
 
 | | Crystal structure of GenB2 in complex with gentamicin X2. | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, ... | | Authors: | Bury, P.S, Araujo, N.C, Oliveira, G.S, Dias, M.V.B. | | Deposit date: | 2024-03-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
6UNW
 
 | | Epoxide hydrolase from an endophytic Streptomyces | | Descriptor: | CACODYLATE ION, CHLORIDE ION, Soluble epoxide hydrolase | | Authors: | Wilson, C, dos Santos, J.C, Dias, M.V.B. | | Deposit date: | 2019-10-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | An epoxide hydrolase from endophytic Streptomyces shows unique structural features and wide biocatalytic activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3BAF
 
 | | Crystal structure of shikimate kinase from Mycobacterium tuberculosis in complex with AMP-PNP | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Shikimate kinase | | Authors: | Faim, L.M, Dias, M.V.B, Vasconcelos, I.G, Basso, L.A, Santos, D.S, Azevedo, W.F, Ruggiero, N.J. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure for shikimate kinase from Mycobacterium tuberculosis in complex with AMP-PNP
To be Published
|
|
8A0Z
 
 | |
7ZZX
 
 | |
8A0N
 
 | |
1PF7
 
 | | CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH IMMUCILLIN H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | De Azevedo Jr, W.F, Canduri, F, Dos Santos, D.M, Pereira, J.H, Dias, M.V.B, Silva, R.G, Mendes, M.A, Palma, M.S, Basso, L.A, Santos, D.S. | | Deposit date: | 2003-05-24 | | Release date: | 2004-06-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for inhibition of human PNP by immucillin-H
Biochem.Biophys.Res.Commun., 309, 2003
|
|
8CRH
 
 | | Crystal structure of Candida auris dihydrofolate reductase complexed with NADPH and cycloguanil | | Descriptor: | 5-(4-chlorophenyl)-6,6-dimethyl-1,4-dihydro-1,3,5-triazine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kirkman, T.J, Dias, M.V.B. | | Deposit date: | 2023-03-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of dihydrofolate reductase from the emerging pathogenic fungus Candida auris.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CQA
 
 | |
8CQ8
 
 | |
8CQ9
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 5-(cyclopropylethynyl)-6-(4-(trifluoromethyl)phenyl)pyrimidine-2,4-diamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(~{E})-2-cyclopropylethenyl]-6-[4-(trifluoromethyl)phenyl]pyrimidine-2,4-diamine, COBALT (II) ION, ... | | Authors: | Kirkman, T.J, Dias, M.V.B, Coyne, A.G. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Expansion of a series of pyrimidine derivatives utilising fragment-based merging leads to increased affinity to Mycobacterium tuberculosis dihydrofolate reductase (unpublished currently)
To Be Published
|
|
7SFP
 
 | |
7RU7
 
 | | Crystal structure of BtrK, a decarboxylase involved in butirosin biosynthesis | | Descriptor: | DI(HYDROXYETHYL)ETHER, L-glutamyl-[BtrI acyl-carrier protein] decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Arenas, L.A.R, Paiva, F.C.R, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BtrK, a decarboxylase involved in the (S)-4-amino-2-hydroxybutyrate (AHBA) formation during butirosin biosynthesis
J.Mol.Struct., 1267, 2022
|
|
7SFN
 
 | |
6D4K
 
 | |
