6RR0
 
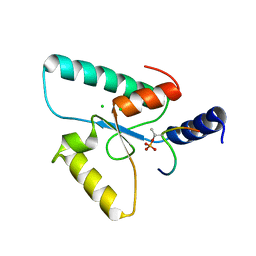 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ubp10 pT123 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6O3C
 
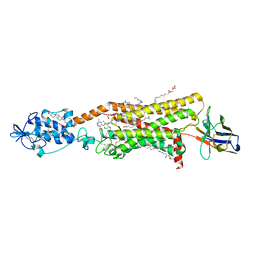 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
6RRV
 
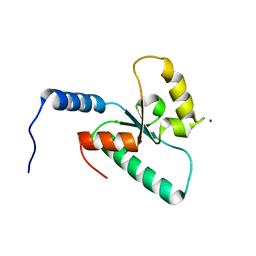 | | Crystal structure of the Sir4 H-BRCT domain | | Descriptor: | BROMIDE ION, CHLORIDE ION, Regulatory protein SIR4 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6QSZ
 
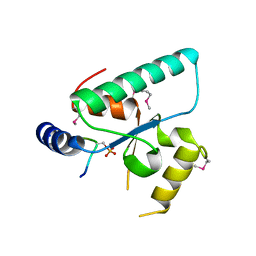 | | Crystal structure of the Sir4 H-BRCT domain in complex with Esc1 pS1450 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Silent chromatin protein ESC1 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5OMB
 
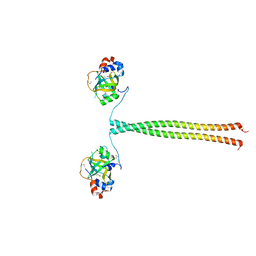 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 N-OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA damage checkpoint protein LCD1, ... | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5OMC
 
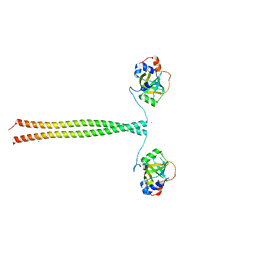 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 (K45E mutant) N-OB domain | | Descriptor: | CHLORIDE ION, DNA damage checkpoint protein LCD1, Replication factor A protein 1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5OMD
 
 | | Crystal structure of S. cerevisiae Ddc2 N-terminal coiled-coil domain | | Descriptor: | DNA damage checkpoint protein LCD1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5M1X
 
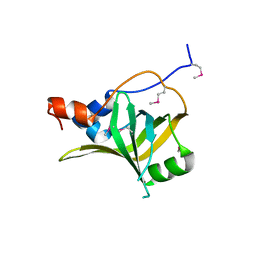 | | Crystal structure of S. cerevisiae Rfa1 N-OB domain mutant (K45E) | | Descriptor: | Replication factor A protein 1 | | Authors: | Seeber, A, Hegnauer, A.M, Hustedt, N, Deshpande, I, Poli, J, Eglinger, J, Pasero, P, Gut, H, Shinohara, M, Hopfner, K.P, Shimada, K, Gasser, S.M. | | Deposit date: | 2016-10-11 | | Release date: | 2016-12-07 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPA Mediates Recruitment of MRX to Forks and Double-Strand Breaks to Hold Sister Chromatids Together.
Mol. Cell, 64, 2016
|
|
6QTM
 
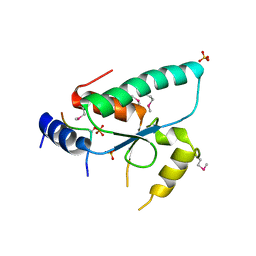 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ty5 pS1095 peptide | | Descriptor: | Regulatory protein SIR4, Ribonuclease H, SULFATE ION | | Authors: | Gut, H, Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-18 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
