2VI7
 
 | | Structure of a Putative Acetyltransferase (PA1377)from Pseudomonas aeruginosa | | Descriptor: | ACETYLTRANSFERASE PA1377, AZIDE ION, GLYCEROL, ... | | Authors: | Davies, A.M, Tata, R, Chauviac, F.X, Sutton, B.J, Brown, P.R. | | Deposit date: | 2007-11-28 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a Putative Acetyltransferase (Pa1377) from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2BL1
 
 | | Crystal structure of a putative phosphinothricin Acetyltransferase (PA4866) from Pseudomonas aeruginosa PAC1 | | Descriptor: | AZIDE ION, GLYCEROL, PUTATIVE PHOSPHINOTHRICIN N-ACETYLTRANSFERASE PA4866, ... | | Authors: | Davies, A.M, Tata, R, Agha, R, Sutton, B.J, Brown, P.R. | | Deposit date: | 2005-02-24 | | Release date: | 2005-09-21 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Putative Phosphinothricin Acetyltransferase (Pa4866) from Pseudomonas Aeruginosa Pac1
Proteins: Struct., Funct., Bioinf., 61, 2005
|
|
5OTJ
 
 | | Monomeric polcalcin (Phl p 7) in complex with two identical allergen-specific antibodies | | Descriptor: | 1,2-ETHANEDIOL, 102.1F10 Fab heavy chain, 102.1F10 Fab light chain, ... | | Authors: | Mitropoulou, A.N, Davies, A.M, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2017-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a patient-derived antibody in complex with allergen reveals simultaneous conventional and superantigen-like recognition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6Y0L
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N, R253G, S254G mutant | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor membrane-bound form, SULFATE ION | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6Y0M
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N mutant | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor membrane-bound form | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6EYO
 
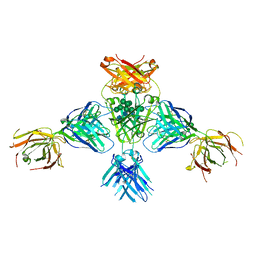 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6EYN
 
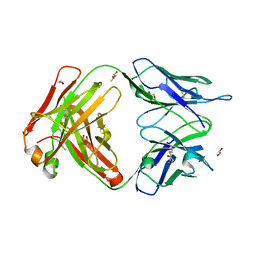 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
5MOI
 
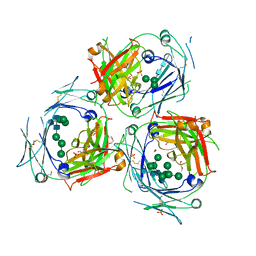 | | Crystal structure of human IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
4LJO
 
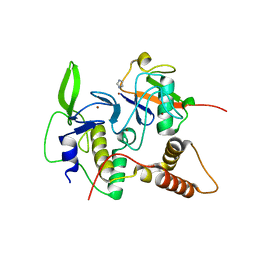 | | Structure of an active ligase (HOIP)/ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, IMIDAZOLE, Polyubiquitin-C, ... | | Authors: | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
5MOJ
 
 | | Crystal structure of IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, MacDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
5MOL
 
 | | Human IgE-Fc crystal structure | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
4LJP
 
 | | Structure of an active ligase (HOIP-H889A)/ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-C, ZINC ION | | Authors: | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
4LJQ
 
 | | Crystal structure of the catalytic core of E3 ligase HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, ZINC ION | | Authors: | Stieglitz, B, Rana, R.R, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
5O9M
 
 | | Crystal structure of human Histamine-Releasing Factor (HRF/TCTP)containing a disulphide-linked dimer | | Descriptor: | DI(HYDROXYETHYL)ETHER, Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
5O9L
 
 | | Crystal structure of human Histamine-Releasing Factor (HRF/TCTP) | | Descriptor: | Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
4AOJ
 
 | | Human TrkA in complex with the inhibitor AZ-23 | | Descriptor: | 5-chloranyl-N2-[(1S)-1-(5-fluoranylpyridin-2-yl)ethyl]-N4-(3-propan-2-yloxy-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR, ZINC ION | | Authors: | Wang, T, Lamb, M.L, Block, M.H, Davies, A.M, Han, Y, Hoffmann, E, Ioannidis, S, Josey, J.A, Liu, Z, Lyne, P.D, MacIntyre, T, Mohr, P.J, Omer, C.A, Sjogren, T, Thress, K, Wang, B, Wang, H, Yu, D, Zhang, H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Disubstituted Imidazo[4,5-B]Pyridines and Purines as Potent Trka Inhibitors
Acs Med.Chem.Lett., 3, 2012
|
|
5MOK
 
 | | Crystal structure of human IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
6EZH
 
 | | Torpedo californica AChE in complex with indolic multi-target directed ligand | | Descriptor: | 1-(7-chloranyl-4-methoxy-1~{H}-indol-5-yl)-3-[1-(cyclohexylmethyl)piperidin-4-yl]propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Santoni, G, Lalut, J, Karila, D, Lecoutey, C, Davis, A, Nachon, F, Sillman, I, Sussman, J, Weik, M, Maurice, T, Dallemagne, P, Rochais, C. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel multitarget-directed ligands targeting acetylcholinesterase and sigma1receptors as lead compounds for treatment of Alzheimer's disease: Synthesis, evaluation, and structural characterization of their complexes with acetylcholinesterase.
Eur J Med Chem, 162, 2018
|
|
5O9K
 
 | | Crystal structure of Murine Histmaine-Releasing Factor (HRF/TCTP) | | Descriptor: | GLYCEROL, Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.014 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
6EZG
 
 | | Torpedo californica AChE in complex with indolic multi-target directed ligand | | Descriptor: | 1-(7-chloranyl-4-methoxy-1~{H}-indol-5-yl)-3-[1-(phenylmethyl)piperidin-4-yl]propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, Lalut, J, Karila, D, Lecoutey, C, Davis, A, Nachon, F, Sillman, I, Sussman, J, Weik, M, Maurice, T, Dallemagne, P, Rochais, C. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel multitarget-directed ligands targeting acetylcholinesterase and sigma1receptors as lead compounds for treatment of Alzheimer's disease: Synthesis, evaluation, and structural characterization of their complexes with acetylcholinesterase.
Eur J Med Chem, 162, 2018
|
|
1RGD
 
 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
6TCN
 
 | | Crystal structure of the omalizumab Fab - crystal form II | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TCP
 
 | | Crystal structure of the omalizumab Fab Leu158Pro light chain mutant - crystal form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Omalizumab Fab Leu158Pro light chain mutant, ... | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TCO
 
 | | Crystal structure of the omalizumab Fab Leu158Pro light chain mutant - crystal form I | | Descriptor: | 1,2-ETHANEDIOL, Omalizumab Fab Leu158Pro light chain mutant, SULFATE ION | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TCS
 
 | | Crystal structure of the omalizumab scFv | | Descriptor: | DI(HYDROXYETHYL)ETHER, Omalizumab scFv | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
