1SDN
 
 | |
6VBC
 
 | |
6VBL
 
 | |
6C4R
 
 | | Staphylopine dehydrogenase (SaODH) - Apo | | Descriptor: | GLYCEROL, SULFATE ION, Staphylopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4T
 
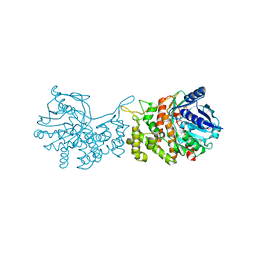 | | Staphylopine dehydrogenase (SaODH) - NADP+ bound | | Descriptor: | GLYCEROL, Staphylopine dehydrogenase, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4L
 
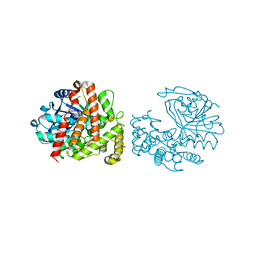 | | Yersinopine dehydrogenase (YpODH) - Apo | | Descriptor: | Yersinopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
1HE8
 
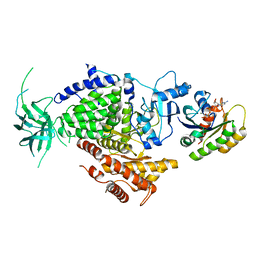 | | Ras G12V - PI 3-kinase gamma complex | | Descriptor: | MAGNESIUM ION, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT, GAMMA ISOFORM, ... | | Authors: | Pacold, M.E, Suire, S, Perisic, O, Lara-Gonzalez, S, Davis, C.T, Hawkins, P.T, Walker, E.H, Stephens, L, Eccleston, J.F, Williams, R.L. | | Deposit date: | 2000-11-20 | | Release date: | 2001-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure and Functional Analysis of Ras Binding to its Effector Phosphoinositide 3-Kinase Gamma
Cell(Cambridge,Mass.), 103, 2000
|
|
1NJ4
 
 | |
1NZO
 
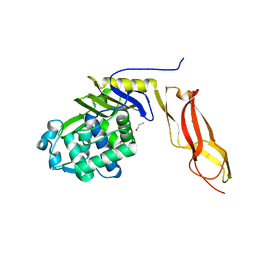 | | The crystal structure of wild type penicillin-binding protein 5 from E. coli | | Descriptor: | BETA-MERCAPTOETHANOL, Penicillin-binding protein 5 | | Authors: | Nicholas, R.A, Krings, S, Tomberg, J, Nicola, G, Davies, C. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of wild-type penicillin-binding protein 5 from Escherichia coli: implications for deacylation of the acyl-enzyme complex.
J.Biol.Chem., 278, 2003
|
|
3MZD
 
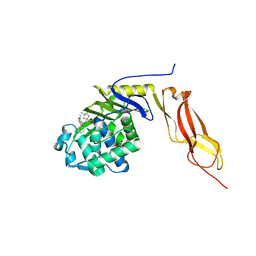 | | Structure of penicillin-binding protein 5 from E. coli: cloxacillin acyl-enzyme complex | | Descriptor: | (2R,4S)-2-[(1S)-1-({[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}amino)-2-oxoethyl]-5,5-dimethyl-1,3-thiazolid ine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3MZE
 
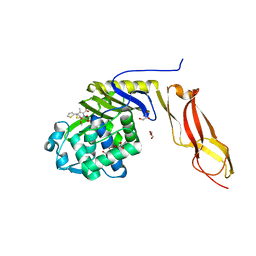 | | Structure of penicillin-binding protein 5 from E.coli: cefoxitin acyl-enzyme complex | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
1DIV
 
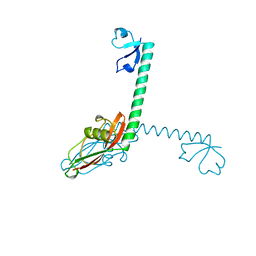 | | RIBOSOMAL PROTEIN L9 | | Descriptor: | RIBOSOMAL PROTEIN L9 | | Authors: | Hoffman, D.W, Cameron, C, Davies, C, Gerchman, S.E, Kycia, J.H, Porter, S, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-07-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein.
EMBO J., 13, 1994
|
|
6P54
 
 | |
6P55
 
 | |
6P56
 
 | |
6P52
 
 | |
3UN7
 
 | |
6P53
 
 | |
5HM6
 
 | |
3LO7
 
 | |
1TZC
 
 | | Crystal structure of phosphoglucose/phosphomannose isomerase from Pyrobaculum aerophilum in complex with 5-phosphoarabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A novel phosphoglucose/phosphomannose isomease from the crenarchaeon Pyrobaculum aerophilum is a member of the PGI superfamily: structural evidence at 1.16 A resolution
J.Biol.Chem., 279, 2004
|
|
1NZU
 
 | |
1TZB
 
 | | Crystal structure of native phosphoglucose/phosphomannose isomerase from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, SULFATE ION, glucose-6-phosphate isomerase, ... | | Authors: | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A novel phosphoglucose/phosphomannose isomease from the crenarchaeon Pyrobaculum aerophilum is a member of the PGI superfamily: structural evidence at 1.16 A resolution
J.Biol.Chem., 279, 2004
|
|
1QX4
 
 | | Structrue of S127P mutant of cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bewley, M.C, Davis, C.A, Marohnic, C.C, Taormina, D, Barber, M.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the S127P mutant of cytochrome b5 reductase that causes methemoglobinemia shows the AMP moiety of the flavin occupying the substrate binding site
Biochemistry, 42, 2003
|
|
1S3I
 
 | |
