2BOH
 
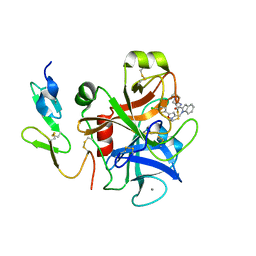 | | Crystal structure of factor Xa in complex with compound "1" | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR XA | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-11 | | Release date: | 2006-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BQ7
 
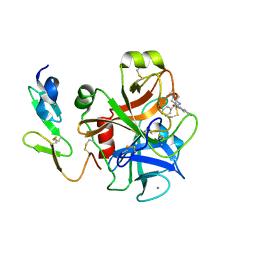 | | Crystal structure of factor Xa in complex with 43 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, FACTOR XA, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2C19
 
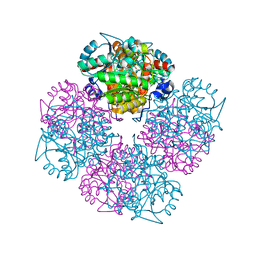 | | 5-(4-Carboxy-2-oxo-butylsulfanyl)-4-oxo-pentanoic acid acid bound to Porphobilinogen synthase from Pseudomonas aeruginosa | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION, SODIUM ION | | Authors: | Frere, F, Nentwich, M, Gacond, S, Heinz, D.W, Neier, R, Frankenberg-Dinkel, N. | | Deposit date: | 2005-09-11 | | Release date: | 2006-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the Active Site of Pseudomonas Aeruginosa Porphobilinogen Synthase Using Newly Developed Inhibitors.
Biochemistry, 45, 2006
|
|
2BMG
 
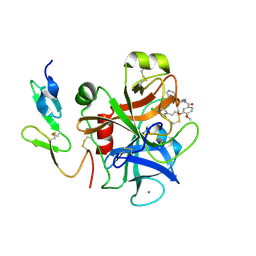 | | Crystal structure of factor Xa in complex with 50 | | Descriptor: | 3-[2-(2,4-DICHLOROPHENYL)ETHOXY]-4-METHOXY-N-[(1-PYRIDIN-4-YLPIPERIDIN-4-YL)METHYL]BENZAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Schreuder, H, Matter, H, Will, D.W, Nazare, M, Laux, V, Wehner, V, Loenze, P, Liesum, A. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Requirements for Factor Xa Inhibition by 3-Oxybenzamides with Neutral P1 Substituents: Combining X-Ray Crystallography, 3D-Qsar and Tailored Scoring Functions
J.Med.Chem., 48, 2005
|
|
2BKB
 
 | | q69e-FeSOD | | Descriptor: | FE (II) ION, SUPEROXIDE DISMUTASE [FE] | | Authors: | Yikilmaz, E, Rodgers, D.W, Miller, A.-F. | | Deposit date: | 2005-02-14 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crucial Importance of Chemistry in the Structure-Function Link: Manipulating Hydrogen Bonding in Iron-Containing Superoxide Dismutase.
Biochemistry, 45, 2006
|
|
5THS
 
 | | Crystal Structure of G302A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
1I90
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-8520 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 4-AMINO-3,4-DIHYDRO-2-(3-METHOXYPROPYL)-, 1,1-DIOXIDE, (R) | | Descriptor: | 4-AMINO-6-[N-(3-METHOXYLPROPYL)-2H-THIENO[3,2-E][1,2]THIAZINE 1,1-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I9O
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
5THU
 
 | | Crystal Structure of G304A HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
2AET
 
 | | R304K trichodiene synthase: Complex with Mg, pyrophosphate, and (4S)-7-azabisabolene | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-07-23 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Role of arginine-304 in the diphosphate-triggered active site closure mechanism of trichodiene synthase.
Biochemistry, 44, 2005
|
|
1ZSA
 
 | | CARBONIC ANHYDRASE II MUTANT E117Q, APO FORM | | Descriptor: | CARBONIC ANHYDRASE II | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1996-01-09 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reversal of the hydrogen bond to zinc ligand histidine-119 dramatically diminishes catalysis and enhances metal equilibration kinetics in carbonic anhydrase II.
Biochemistry, 35, 1996
|
|
1ZSB
 
 | | CARBONIC ANHYDRASE II MUTANT E117Q, TRANSITION STATE ANALOGUE ACETAZOLAMIDE | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1996-01-09 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reversal of the hydrogen bond to zinc ligand histidine-119 dramatically diminishes catalysis and enhances metal equilibration kinetics in carbonic anhydrase II.
Biochemistry, 35, 1996
|
|
1ZSC
 
 | | CARBONIC ANHYDRASE II MUTANT E117Q, HOLO FORM | | Descriptor: | CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1996-01-09 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversal of the hydrogen bond to zinc ligand histidine-119 dramatically diminishes catalysis and enhances metal equilibration kinetics in carbonic anhydrase II.
Biochemistry, 35, 1996
|
|
1G1D
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2-FLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2-FLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-11 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Contribution of Fluorine to Protein-Ligand Affinity in the Binding of Fluoroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1G4O
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-PHENYLMETHYLBENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-BENZYL-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1HOZ
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX | | Descriptor: | CALCIUM ION, GLYCEROL, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1HQF
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
1HM4
 
 | |
1HQ8
 
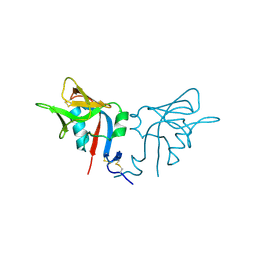 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | Descriptor: | NKG2-D | | Authors: | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
1HQG
 
 | | CRYSTAL STRUCTURE OF THE H141C ARGINASE VARIANT COMPLEXED WITH PRODUCTS ORNITHINE AND UREA | | Descriptor: | ARGINASE 1, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
6UFN
 
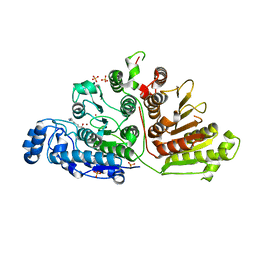 | |
6UHV
 
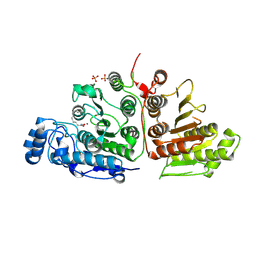 | |
6UII
 
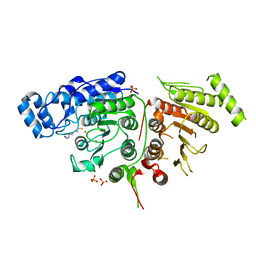 | |
2C18
 
 | | 5-(4-Carboxy-2-oxo-butane-1-sulfonyl)-4-oxo-pentanoic acid bound to Porphobilinogen synthase from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Frere, F, Nentwich, M, Gacond, S, Heinz, D.W, Neier, R, Frankenberg-Dinkel, N. | | Deposit date: | 2005-09-11 | | Release date: | 2006-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the Active Site of Pseudomonas Aeruginosa Porphobilinogen Synthase Using Newly Developed Inhibitors.
Biochemistry, 45, 2006
|
|
2C13
 
 | | 5-hydroxy-levulinic acid bound to Porphobilinogen synthase from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Frere, F, Nentwich, M, Gacond, S, Heinz, D.W, Neier, R, Frankenberg-Dinkel, N. | | Deposit date: | 2005-09-11 | | Release date: | 2006-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the Active Site of Pseudomonas Aeruginosa Porphobilinogen Synthase Using Newly Developed Inhibitors.
Biochemistry, 45, 2006
|
|
