2VGY
 
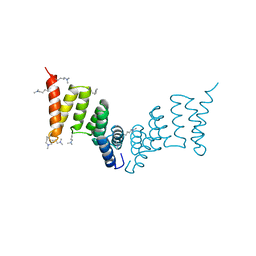 | | Crystal structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD (alternative dimer) | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
7CDH
 
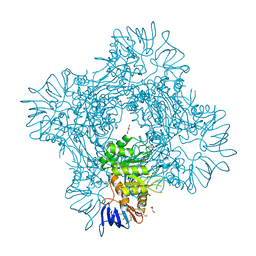 | | Crystal structure of Betaaspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum-AW-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isoaspartyl dipeptidase, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
7CV2
 
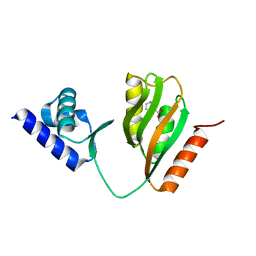 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | Descriptor: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
2V8K
 
 | |
4BFB
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, XA4813 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2V78
 
 | | Crystal structure of Sulfolobus solfataricus 2-keto-3-deoxygluconate kinase | | Descriptor: | FRUCTOKINASE | | Authors: | Potter, J.A, Theodossis, A, Kerou, M, Lamble, H.J, Bull, S.D, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Sulfolobus Solfataricus 2-Keto-3-Deoxygluconate Kinase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2V4V
 
 | | Crystal Structure of a Family 6 Carbohydrate-Binding Module from Clostridium cellulolyticum in complex with xylose | | Descriptor: | GH59 GALACTOSIDASE, SODIUM ION, beta-D-xylopyranose | | Authors: | Abbott, D.W, Ficko-Blean, E, Lammerts van Bueren, A, Coutinho, P.M, Henrissat, B, Gilbert, H.J, Boraston, A.B. | | Deposit date: | 2008-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the Structural and Functional Diversity of Plant Cell Wall Specific Family 6 Carbohydrate Binding Modules.
Biochemistry, 48, 2009
|
|
2V5G
 
 | | Crystal structure of the mutated N263A YscU C-terminal domain | | Descriptor: | CHLORIDE ION, YSCU | | Authors: | Wiesand, U, Sorg, I, Amstutz, M, Wagner, S, Van Den Heuvel, J, Luehrs, T, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Type III Secretion Recognition Protein Yscu from Yersinia Enterocolitica
J.Mol.Biol., 385, 2009
|
|
2VNG
 
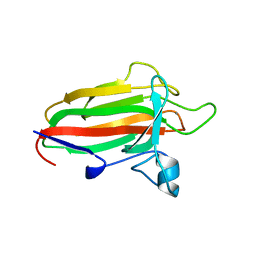 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens in complex with blood group A-trisaccharide ligand. | | Descriptor: | CALCIUM ION, CPE0329, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
4D3Z
 
 | |
4D8I
 
 | |
4A7I
 
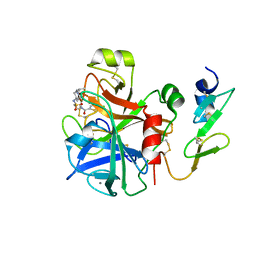 | | Factor Xa in complex with a potent 2-amino-ethane sulfonamide inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID [2-(1--ISOPROPYL-PIPERIDIN-4-YLSULFAMOYL)-ETHYL]-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN XA, CALCIUM ION, ... | | Authors: | Nazare, M, Matter, H, Will, D.W, Wagner, M, Urmann, M, Czech, J, Schreuder, H, Bauer, A, Ritter, K, Wehner, V. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment Deconstruction of Small, Potent Factor Xa Inhibitors: Exploring the Superadditivity Energetics of Fragment Linking in Protein-Ligand Complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1L0I
 
 | | Crystal structure of butyryl-ACP I62M mutant | | Descriptor: | Acyl carrier protein, CACODYLATE ION, SODIUM ION, ... | | Authors: | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2002-02-11 | | Release date: | 2003-02-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray Crystallographic Studies on Butyryl-ACP Reveal Flexibility of the Structure around a Putative Acyl Chain Binding Site
Structure, 10, 2002
|
|
1SAY
 
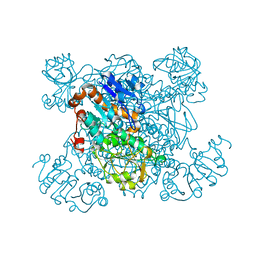 | | L-ALANINE DEHYDROGENASE COMPLEXED WITH PYRUVATE | | Descriptor: | L-ALANINE DEHYDROGENASE, PYRUVIC ACID | | Authors: | Baker, P.J, Sawa, Y, Shibata, H, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase.
Nat.Struct.Biol., 5, 1998
|
|
4A1T
 
 | | Co-Complex of the of NS3-4A protease with the inhibitory peptide CP5- 46-A (in-House data) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
8U3Y
 
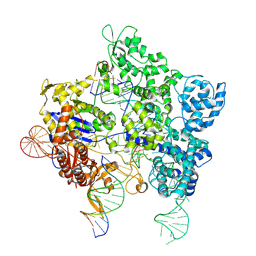 | | SpG Cas9 with NGG PAM DNA target | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
4A10
 
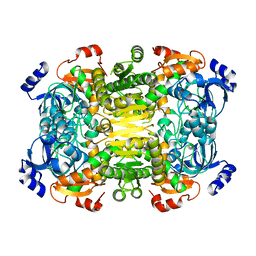 | | Apo-structure of 2-octenoyl-CoA carboxylase reductase CinF from streptomyces sp. | | Descriptor: | OCTENOYL-COA REDUCTASE/CARBOXYLASE | | Authors: | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Unusual Carbon Fixation Giving Rise to Diverse Polyketide Extender Units
Nat.Chem.Biol., 8, 2011
|
|
8TZZ
 
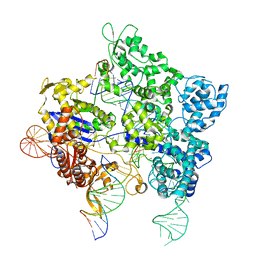 | | SpG Cas9 with NGC PAM DNA target | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | Deposit date: | 2023-08-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
1S9E
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
2RLA
 
 | |
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
4CMP
 
 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
1PS1
 
 | | PENTALENENE SYNTHASE | | Descriptor: | PENTALENENE SYNTHASE, TRIMETHYL LEAD ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-03-23 | | Release date: | 1998-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology.
Science, 277, 1997
|
|
4D3X
 
 | |
7CF6
 
 | | Crystal structure of Beta-aspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum AW-1 in complex with beta-Asp-Leu dipeptide | | Descriptor: | (2S)-2-[[(3S)-3-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-methyl-pentanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
