4DOV
 
 | | Structure of free mouse ORC1 BAH domain | | Descriptor: | Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
4DB4
 
 | | Mss116p DEAD-box helicase domain 2 bound to a chimaeric RNA-DNA duplex | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*G)-D(P*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase MSS116, mitochondrial | | Authors: | Mallam, A.L, Del Campo, M, Gilman, B.D, Sidote, D.J, Lambowitz, A. | | Deposit date: | 2012-01-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p.
Nature, 490, 2012
|
|
4DC6
 
 | | Crystal Structure of Thaumatin Exposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Mulichak, A.M, Becker, M, Kissick, D.J, Keefe, L.J, Fischetti, R.F, Simpson, G.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4DSY
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC24201 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-phenylpyridine-3-carboxylic acid, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
4DC7
 
 | | Crystal Structure of Myoglobin Exposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Becker, M, Mulichak, A.M, Kissick, D.J, Fischetti, R.F, Keefe, L.J, Simpson, G.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1RUZ
 
 | | 1918 H1 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Skehel, J.J, Gamblin, S.J, Haire, L.F, Russell, R.J, Stevens, D.J, Xiao, B, Ha, Y, Vasisht, N, Steinhauer, D.A, Daniels, R.S. | | Deposit date: | 2003-12-12 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and receptor binding properties of the 1918 influenza hemagglutinin.
Science, 303, 2004
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
1Q8L
 
 | | Second Metal Binding Domain of the Menkes ATPase | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Jones, C.E, Daly, N.L, Cobine, P.A, Craik, D.J, Dameron, C.T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and metal binding studies of the second copper binding domain of the Menkes ATPase.
J.Struct.Biol., 143, 2003
|
|
4DVE
 
 | | Crystal structure at 2.1 A of the S-component for biotin from an ECF-type ABC transporter | | Descriptor: | BIOTIN, Biotin transporter BioY, nonyl beta-D-glucopyranoside | | Authors: | Berntsson, R.P.-A, ter Beek, J, Majsnerowska, M, Duurkens, R, Puri, P, Poolman, B, Slotboom, D.J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural divergence of paralogous S components from ECF-type ABC transporters.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6BL9
 
 | |
1QIC
 
 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
1QO8
 
 | | The structure of the open conformation of a flavocytochrome c3 fumarate reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C3 FUMARATE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bamford, V, Dobbin, P.S, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 1999-11-04 | | Release date: | 2000-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Open Conformation of a Flavocytochrome C3 Fumarate Reductase.
Nat.Struct.Biol., 6, 1999
|
|
6ASR
 
 | | REV1 UBM2 domain complex with ubiquitin | | Descriptor: | DNA repair protein REV1, NICKEL (II) ION, Ubiquitin | | Authors: | Miller, D.J. | | Deposit date: | 2017-08-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structures of REV1 UBM2 Domain Complex with Ubiquitin and with a Small-Molecule that Inhibits the REV1 UBM2-Ubiquitin Interaction.
J. Mol. Biol., 430, 2018
|
|
6AVI
 
 | |
4FER
 
 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellohexaose | | Descriptor: | ACETIC ACID, Expansin-yoaJ, GLYCEROL, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-05-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1PX8
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
4EZ4
 
 | | free KDM6B structure | | Descriptor: | Lysine-specific demethylase 6B, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Cheng, Z.J, Patel, D.J. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Nature, 488, 2012
|
|
4F9J
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
4F9D
 
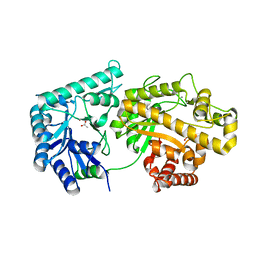 | | Structure of Escherichia coli PgaB 42-655 in complex with nickel | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
4F1N
 
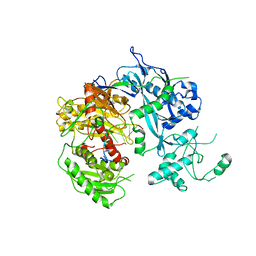 | | Crystal structure of Kluyveromyces polysporus Argonaute with a guide RNA | | Descriptor: | KpAGO, RNA 5'-R(P*UP*AP*AP*AP*AP*AP*AP*AP*A)-3' | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure of yeast Argonaute with guide RNA.
Nature, 486, 2012
|
|
6NIZ
 
 | |
1PKL
 
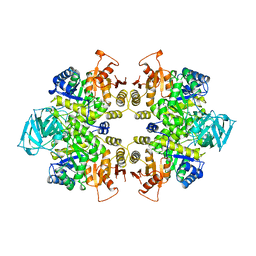 | | THE STRUCTURE OF LEISHMANIA PYRUVATE KINASE | | Descriptor: | PROTEIN (PYRUVATE KINASE), SULFATE ION | | Authors: | Rigden, D.J, Phillips, S.E.V, Michels, P.A.M, Fothergill-Gilmore, L.A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of pyruvate kinase from Leishmania mexicana reveals details of the allosteric transition and unusual effector specificity.
J.Mol.Biol., 291, 1999
|
|
6C1I
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with T0070907 | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Fuhrmann, J, Brust, R, Kojetin, D.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A structural mechanism for directing corepressor-selective inverse agonism of PPAR gamma.
Nat Commun, 9, 2018
|
|
4EB0
 
 | | Crystal structure of Leaf-branch compost bacterial cutinase homolog | | Descriptor: | LCC, THIOCYANATE ION | | Authors: | Sulaiman, S, You, D.J, Eiko, K, Koga, Y, Kanaya, S. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Leaf-branch compost bacterial cutinase homolog
To be Published
|
|
6CSZ
 
 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R3) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | Descriptor: | PaDBS1R3 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
