3ZQ3
 
 | | Crystal Structure of Rat Odorant Binding Protein 3 (OBP3) | | Descriptor: | OBP3 PROTEIN | | Authors: | Portman, K.L, Long, J, Carr, S, Brand, L, Winzor, D.J, Searle, M, Scott, D.J. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enthalpy/Entropy Compensation Effects from Cavity Desolvation Underpin Broad Ligand Binding Selectivity for Rat Odorant Binding Protein 3
Biochemistry, 53, 2014
|
|
2AK0
 
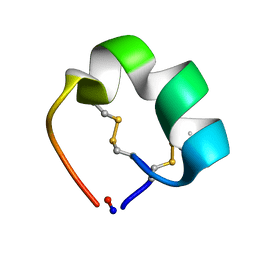 | | Structure of cyclic conotoxin MII-7 | | Descriptor: | Alpha-conotoxin MII | | Authors: | Clark, R.J, Fischer, H, Dempster, L, Daly, N.L, Rosengren, K.J, Nevin, S.T, Meunier, F.A, Adams, D.J, Craik, D.J. | | Deposit date: | 2005-08-02 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the {alpha}-conotoxin MII.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8J
 
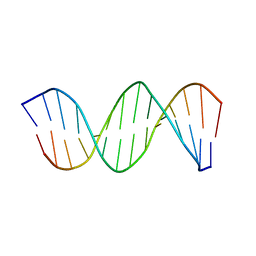 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
4XHK
 
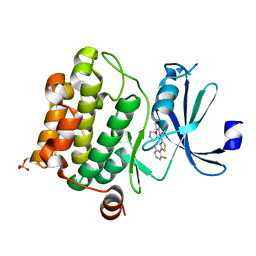 | | PIM1 kinase in complex with Compound 1s | | Descriptor: | 2-(2,6-difluorophenyl)-N-{4-[(3S)-pyrrolidin-3-yloxy]pyridin-3-yl}-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Marcotte, D.J, Silvian, L.F. | | Deposit date: | 2015-01-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of low-nanomolar PIM kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8BAI
 
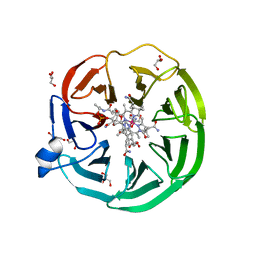 | | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin. | | Descriptor: | CYANOCOBALAMIN, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J, Martinez Felices, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin.
To Be Published
|
|
8BB0
 
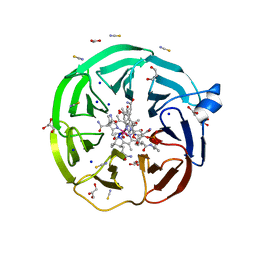 | | The surface-exposed lipo-protein of BtuG2 in complex with hydroxycobalamin. | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J, Felices Martinez, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The surface-exposed lipo-protein of BtuG2 in complex with hydroxycobinamide.
To Be Published
|
|
8BBZ
 
 | |
8BAZ
 
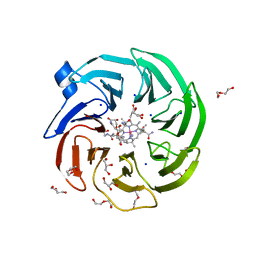 | | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobalamin. | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittaker, J, Martinez-Felices, J.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The surface-exposed lipo-protein of BtuG2 in complex with cyanocobinamide.
To Be Published
|
|
8BHU
 
 | | Crystal Structure of SilF (Ag(I) form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SILVER ION, SULFATE ION, ... | | Authors: | Lithgo, R.M, Carr, S.B, Quigley, A.M, Scott, D.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of SilF (apo form)
To Be Published
|
|
1KJ6
 
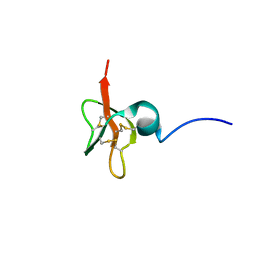 | | Solution Structure of Human beta-Defensin 3 | | Descriptor: | Beta-defensin 3 | | Authors: | Schibli, D.J, Hunter, H.N, Aseyev, V, Starner, T.D, Wiencek, J.M, McCray Jr, P.B, Tack, B.F, Vogel, H.J. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
J.Biol.Chem., 277, 2002
|
|
1KKQ
 
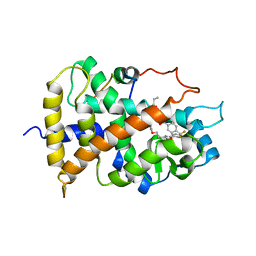 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
8C6D
 
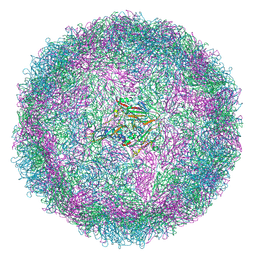 | | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate. | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, Genome polyprotein, Genome polyprotein (Fragment) | | Authors: | Kingston, N.J, Snowden, J.S, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.
Biorxiv, 2023
|
|
1KJ5
 
 | | Solution Structure of Human beta-defensin 1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Schibli, D.J, Hunter, H.N, Aseyev, V, Starner, T.D, Wiencek, J.M, McCray Jr, P.B, Tack, B.F, Vogel, H.J. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
J.Biol.Chem., 277, 2002
|
|
5K1R
 
 | |
1MXN
 
 | | Solution structure of alpha-conotoxin AuIB | | Descriptor: | alpha-conotoxin AuIB | | Authors: | Dutton, J.L, Bansal, P.S, Hogg, R.C, Adams, D.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A New Level of Conotoxin Diversity, a Non-native Disulfide Bond Connectivity in alpha -Conotoxin AuIB Reduces Structural Definition but Increases Biological Activity.
J.Biol.Chem., 277, 2002
|
|
7M3N
 
 | |
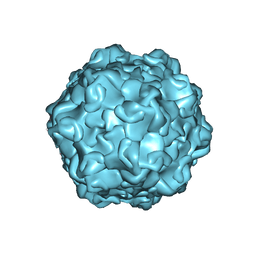
7M3M
 
 | |
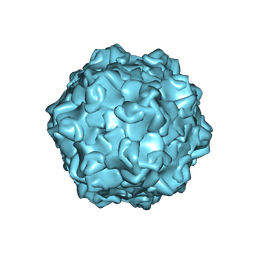
7M3O
 
 | |
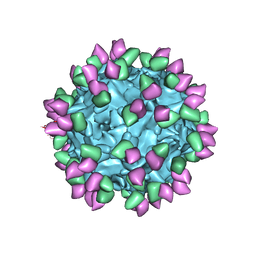
7M3L
 
 | |
4ZJW
 
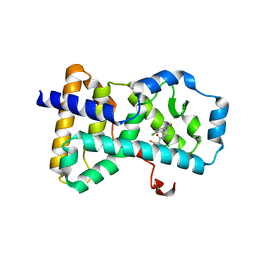 | | RORgamma in complex with inverse agonist 16 | | Descriptor: | 4-chloro-3-[1-(2-chloro-6-fluorobenzoyl)-1,2,3,4-tetrahydroquinolin-6-yl]-N-methylbenzamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.J. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of biaryl carboxylamides as potent ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4ZJR
 
 | | RORgamma in complex with inverse agonist 48 | | Descriptor: | 6-chloro-4'-[(2-chloro-6-fluorobenzoyl)(methyl)amino]-3'-(2,2,2-trifluoroethoxy)biphenyl-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.J. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Discovery of biaryl carboxylamides as potent ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4ZOM
 
 | | RORgamma in complex with inverse agonist 4j. | | Descriptor: | N-{4-[3-(acetylamino)-1-(propan-2-yl)-1H-pyrazol-5-yl]-2-[(1R,5S)-3-azabicyclo[3.1.0]hex-3-yl]phenyl}-2-chloro-6-fluoro-N-methylbenzamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of novel pyrazole-containing benzamides as potent ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4ZSG
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chlorophenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J, Ogg, D.J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
