3QHY
 
 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase, Beta-lactamase inhibitory protein II | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
3QL3
 
 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
3Q44
 
 | |
3QL0
 
 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
3RTR
 
 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SDY
 
 | |
3QHF
 
 | |
3QI0
 
 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase inhibitory protein II, SULFATE ION | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
3QHZ
 
 | | Crystal Structure of human anti-influenza Fab 2D1 | | Descriptor: | Human monoclonal antibody del2D1, Fab Heavy Chain, Fab Light Chain | | Authors: | Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2011-01-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody.
MBio, 2, 2011
|
|
3TC1
 
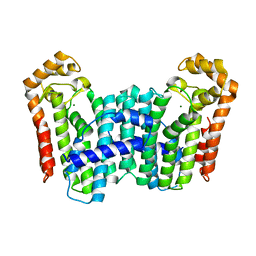 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori | | Descriptor: | MAGNESIUM ION, Octaprenyl Pyrophosphate Synthase | | Authors: | Zhang, J.Y, Zhang, X.L, Li, D.F, Zou, Q.M, Wang, D.C. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori
To be Published
|
|
3Q43
 
 | |
3TDI
 
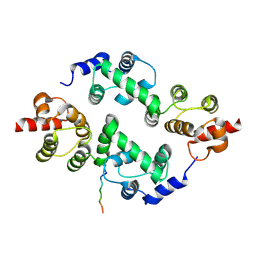 | | yeast Cul1WHB-Dcn1P acetylated Ubc12N complex | | Descriptor: | Defective in cullin neddylation protein 1, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3TDZ
 
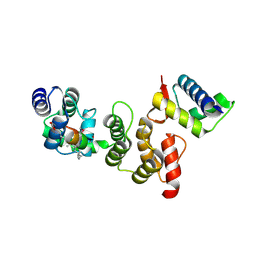 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-stapled acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, STAPLED PEPTIDE | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3TDU
 
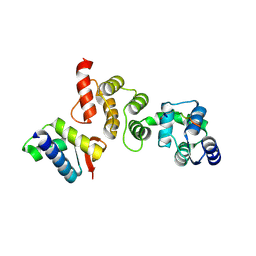 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, NEDD8-conjugating enzyme Ubc12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3R2X
 
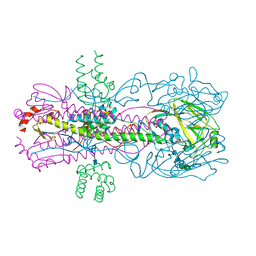 | |
3SXL
 
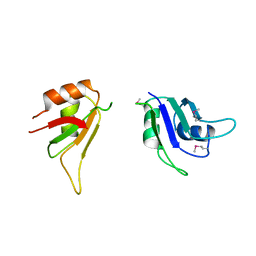 | |
3TG0
 
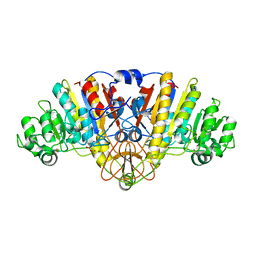 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
3SLK
 
 | |
3U1N
 
 | | Structure of the catalytic core of human SAMHD1 | | Descriptor: | PHOSPHATE ION, SAM domain and HD domain-containing protein 1, ZINC ION | | Authors: | Goldstone, D.C, Ennis-Adeniran, V, Walker, P.A, Haire, L.F, Webb, M, Taylor, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase
Nature, 480, 2011
|
|
3TUI
 
 | |
3TUJ
 
 | |
3TUZ
 
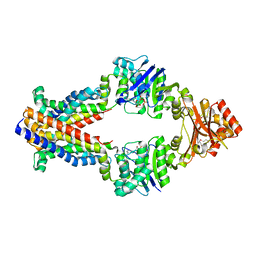 | | Inward facing conformations of the MetNI methionine ABC transporter: CY5 SeMet soak crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-methionine transport system permease protein metI, Methionine import ATP-binding protein MetN, ... | | Authors: | Johnson, E, Nguyen, P, Rees, D.C. | | Deposit date: | 2011-09-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition.
Protein Sci., 21, 2012
|
|
3TMM
 
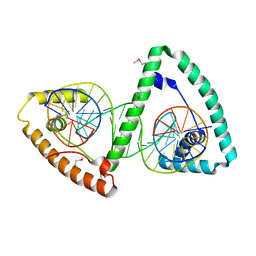 | | TFAM imposes a U-turn on mitochondrial DNA | | Descriptor: | DNA (28-MER), Transcription factor A, mitochondrial | | Authors: | Ngo, H.B, Kaiser, J.T, Chan, D.C. | | Deposit date: | 2011-08-31 | | Release date: | 2011-11-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3LFK
 
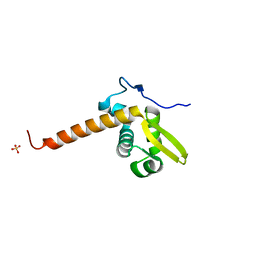 | | A reported archaeal mechanosensitive channel is a structural homolog of MarR-like transcriptional regulators | | Descriptor: | CITRIC ACID, MarR Like Protein, TVG0766549, ... | | Authors: | Liu, Z, Walton, T.A, Rees, D.C. | | Deposit date: | 2010-01-17 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A reported archaeal mechanosensitive channel is a structural homolog of MarR-like transcriptional regulators.
Protein Sci., 19, 2010
|
|
3LZF
 
 | |
