5U5N
 
 | |
2CLR
 
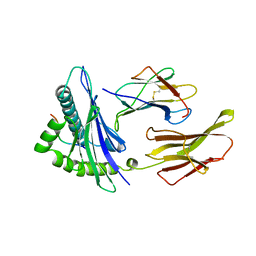 | |
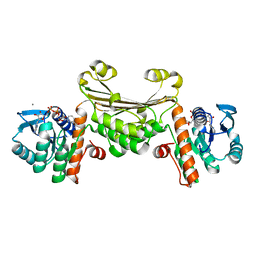
4WZB
 
 | | Crystal Structure of MgAMPPCP-bound Av2-Av1 complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nitrogenase complexes: multiple docking sites for a nucleotide switch protein.
Science, 309, 2005
|
|
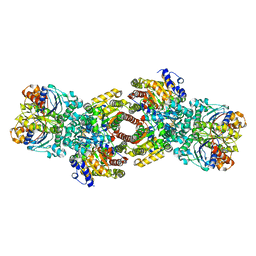
6YR8
 
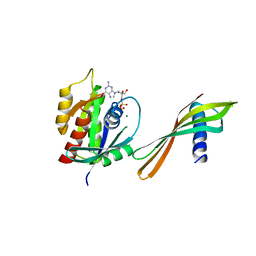 | | Affimer K6 - KRAS protein complex | | Descriptor: | Affimer K6, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
6YXW
 
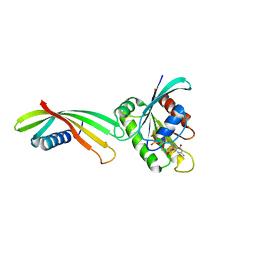 | | Affimer K3 - KRAS protein complex | | Descriptor: | Affimer K3, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
1QRE
 
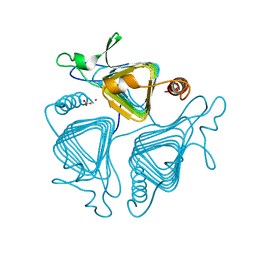 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRL
 
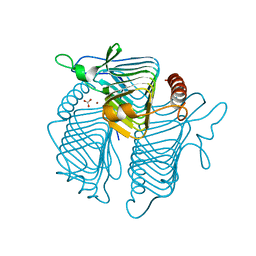 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
5MTF
 
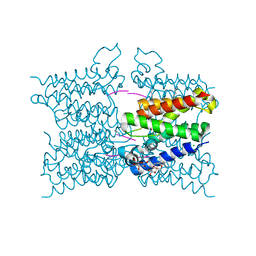 | | A modular route to novel potent and selective inhibitors of rhomboid intramembrane proteases | | Descriptor: | CHLORIDE ION, Rhomboid protease GlpG, inhibitor, ... | | Authors: | Ticha, A, Stanchev, S, Vinothkumar, K.R, Mikles, D.C, Pachl, P, Svehlova, K, Nguyen, M.T.N, Verhelst, S.H.L, Johnson, D, Bachovchin, D, Lepsik, M, Majer, P, Strisovsky, K. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | General and Modular Strategy for Designing Potent, Selective, and Pharmacologically Compliant Inhibitors of Rhomboid Proteases.
Cell Chem Biol, 24, 2017
|
|
1QRF
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION, SULFATE ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRM
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, SULFATE ION, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QQD
 
 | |
1QPG
 
 | | 3-PHOSPHOGLYCERATE KINASE, MUTATION R65Q | | Descriptor: | 3-PHOSPHOGLYCERATE KINASE, 3-PHOSPHOGLYCERIC ACID, MAGNESIUM-5'-ADENYLY-IMIDO-TRIPHOSPHATE | | Authors: | Mcphillips, T.M, Hsu, B.T, Sherman, M.A, Mas, M.T, Rees, D.C. | | Deposit date: | 1996-01-04 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the R65Q mutant of yeast 3-phosphoglycerate kinase complexed with Mg-AMP-PNP and 3-phospho-D-glycerate.
Biochemistry, 35, 1996
|
|
1QQ0
 
 | | COBALT SUBSTITUTED CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-10 | | Release date: | 1999-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
6VPL
 
 | |
6WA8
 
 | |
6VPH
 
 | |
6VQU
 
 | | Structure of a bacterial Atm1-family ABC exporter | | Descriptor: | ATM1-type heavy metal exporter | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A structural framework for unidirectional transport by a bacterial ABC exporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VPJ
 
 | |
5BNR
 
 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5AO6
 
 | | Endo180 D1-4, trigonal form | | Descriptor: | C-TYPE MANNOSE RECEPTOR 2 | | Authors: | Paracuellos, P, Briggs, D.C, Carafoli, F, Loncar, T, Hohenester, E. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Insights Into Collagen Uptake by C-Type Mannose Receptors from the Crystal Structure of Endo180 Domains 1-4.
Structure, 23, 2015
|
|
5BQS
 
 | | S. Pneumoniae Fabh with small molecule inhibitor 4 | | Descriptor: | 1-{5-[2-chloro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SODIUM ION | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-29 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5BNS
 
 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}-N-(quinolin-6-ylmethyl)piperidine-4-carboxamide, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5C7C
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 18 | | Descriptor: | (2R)-4-[2-(6-chloro-3,3-dimethyl-2,3-dihydro-1H-indol-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
5LYZ
 
 | |
4M6K
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
