4CV9
 
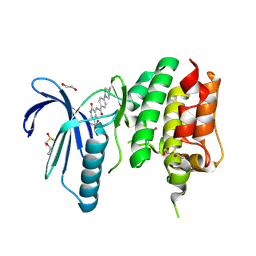 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-Methylpiperazin-1-yl)-N-(2-oxo-5-(pyridin-4-yl)-1,2-dihydropyridin-3-yl)benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
4B1B
 
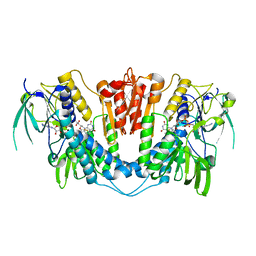 | | Crystal structure of Plasmodium falciparum oxidised Thioredoxin Reductase at 2.9 angstrom | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE | | Authors: | Boumis, G, Giardina, G, Dimastrogiovanni, D, Angelucci, F, Saccoccia, F, Brunori, M, Bellelli, A, Miele, A.E. | | Deposit date: | 2012-07-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Plasmodium Falciparum Thioredoxin Reductase, a Validated Drug Target.
Biochem.Biophys.Res.Commun., 425, 2012
|
|
4B2X
 
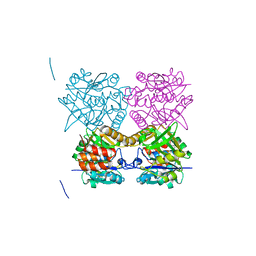 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-butyl-5-(cyclopentylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B4M
 
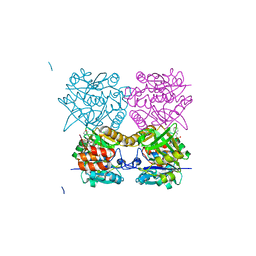 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4FZN
 
 | |
4BBJ
 
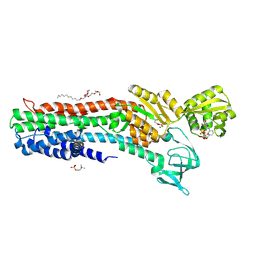 | | Copper-transporting PIB-ATPase in complex with beryllium fluoride representing the E2P state | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mattle, D, Gourdon, P, Nissen, P. | | Deposit date: | 2012-09-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Copper-Transporting P-Type Atpases Use a Unique Ion-Release Pathway
Nat.Struct.Mol.Biol., 21, 2014
|
|
4BFC
 
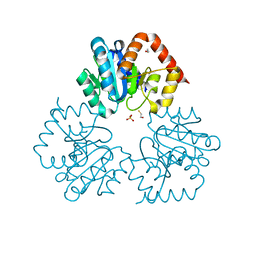 | | Crystal structure of the C-terminal CMP-Kdo binding domain of WaaA from Acinetobacter baumannii | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONIC-ACID TRANSFERASE, BETA-MERCAPTOETHANOL, SULFATE ION | | Authors: | Kimbung, Y.R, Hakansson, M, Logan, D, Wang, P.F, Schulz, M, Mamat, U, Woodard, R.W. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the C-Terminal Cmp-Kdo Binding Domain of Waaa from Acinetobacter Baumannii
To be Published
|
|
4ARK
 
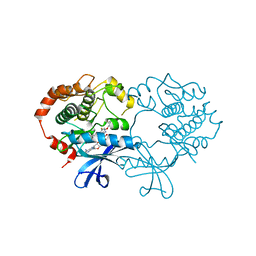 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN MAP KINASE KINASE 1 (MEK1) IN COMPLEX WITH A SMALL MOLECULE INHIBITOR AND ADP | | Descriptor: | 2-([3R-3,4-dihydroxybutyl]oxy)-4-fluoro-6-[(2-fluoro-4-iodophenyl)amino]benzamide, ADENOSINE-5'-DIPHOSPHATE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, ... | | Authors: | Hartung, I.V, Hitchcock, M, Puehler, F, Neuhaus, R, Scholz, A, Hammer, S, Petersen, K, Siemeister, G, Brittain, D, Hillig, R.C. | | Deposit date: | 2012-04-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Allosteric Mek Inhibitors - Part 1: Venturing Into Unexplored Sar Territories
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ASY
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4AZQ
 
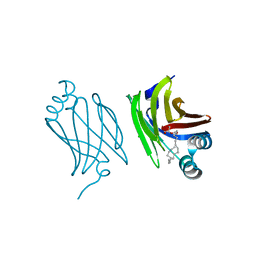 | | Murine epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid 2-arachidonoylglycerol | | Descriptor: | 2-hydroxy-1-(hydroxymethyl)ethyl icosanoate, CHLORIDE ION, FATTY ACID-BINDING PROTEIN, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4GK1
 
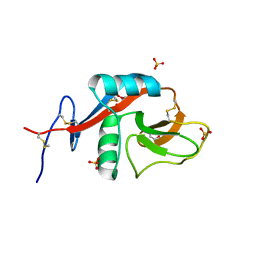 | | Crystal structure of CD23 lectin domain mutant D270A | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
4AZO
 
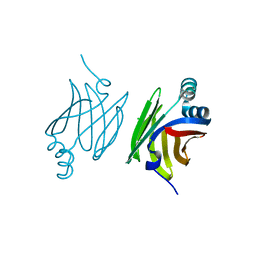 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag removed | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BP9
 
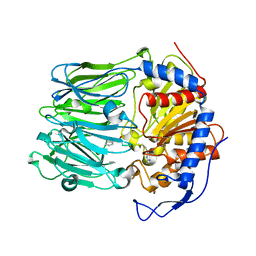 | | Oligopeptidase B from Trypanosoma brucei with covalently bound antipain - closed form | | Descriptor: | ANTIPAIN, OLIGOPEPTIDASSE B | | Authors: | Canning, P, Rea, D, Morty, R, Fulop, V. | | Deposit date: | 2013-05-23 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of Trypanosoma Brucei Oligopeptidase B Broaden the Paradigm of Catalytic Regulation in Prolyl Oligopeptidase Family Enzymes.
Plos One, 8, 2013
|
|
4BMW
 
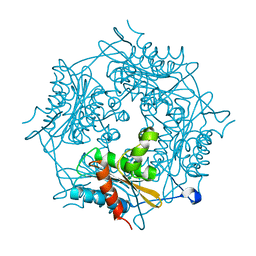 | | Crystal structure of the Streptomyces reticuli HbpS E78D, E81D double mutant | | Descriptor: | EXTRACELLULAR HAEM-BINDING PROTEIN | | Authors: | Wagener, S, Kursula, I, Wedderhoff, I, Groves, M.R, Ortiz de Orue Lucana, D. | | Deposit date: | 2013-05-11 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Iron Binding at Specific Sites within the Octameric Hbps Protects Streptomycetes from Iron-Mediated Oxidative Stress.
Plos One, 8, 2013
|
|
4FZM
 
 | | Crystal structure of the bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION | | Authors: | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
4FZS
 
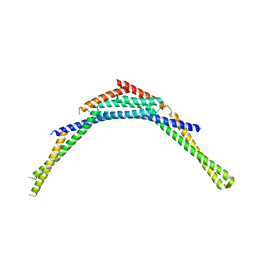 | | Structure of human SNX1 BAR domain | | Descriptor: | Sorting nexin-1 | | Authors: | van Weering, J.R.T, Sessions, R.B, Traer, C.J, Kloer, D.P, Bhatia, V.K, Stamou, D, Hurley, J.H, Cullen, P.J. | | Deposit date: | 2012-07-07 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insight into vesicle-to-tubule membrane remodeling by SNX-BAR proteins
To be Published
|
|
4BOS
 
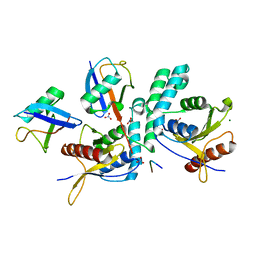 | | Structure of OTUD2 OTU domain in complex with Ubiquitin K11-linked peptide | | Descriptor: | MAGNESIUM ION, NITRATE ION, OTUD2, ... | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
4G21
 
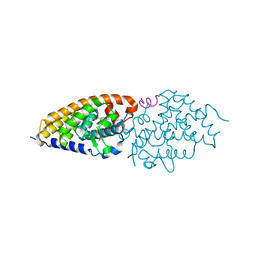 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G2I
 
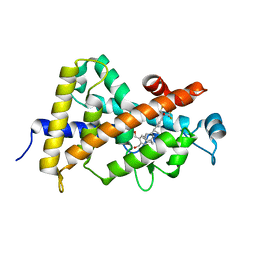 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Vitamin D3 receptor | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4BT4
 
 | | acetolactate decarboxylase with a bound (2S,3S)-2,3-Dihydroxy-2- methylbutanoic acid | | Descriptor: | (2S,3S)-2,3-dihydroxy-2-methylbutanoic acid, ALPHA-ACETOLACTATE DECARBOXYLASE, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase.
Acs Chem.Biol., 8, 2013
|
|
4BS0
 
 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
4BWY
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI8 (R32) | | Descriptor: | P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4BOZ
 
 | | Structure of OTUD2 OTU domain in complex with K11-linked di ubiquitin | | Descriptor: | GLYCEROL, UBIQUITIN THIOESTERASE OTU1, UBIQUITIN-C | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
4GKO
 
 | | Crystal structure of the calcium2+-bound human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 | | Descriptor: | CALCIUM ION, Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, ... | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
