5TSD
 
 | | Crystal structure of NADPH-dependent 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH and oxalate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALIC ACID, Probable hydroxyacid dehydrogenase protein | | Authors: | Matelska, D, Shabalin, I.G, Kutner, J, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of NADPH-dependent 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH and oxalate
to be published
|
|
3W0S
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, MAGNESIUM ION, ... | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
2J74
 
 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, YVFO, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-10-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
3W0M
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hygromycin-B 4-O-kinase | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
2J8L
 
 | | FXI Apple 4 domain loop-out conformation | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5TUN
 
 | | Crystal structure of uninhibited human Cathepsin K at 1.62 Angstrom resolution | | Descriptor: | Cathepsin K | | Authors: | Aguda, A.H, Kruglyak, N, Nguyen, N.T, Law, S, Bromme, D, Brayer, G.D. | | Deposit date: | 2016-11-06 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib.
Biochem. J., 474, 2017
|
|
7YNF
 
 | | CCA-bound alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, copper;trisodium;18-(2-carboxylatoethyl)-20-(carboxylatomethyl)-12-ethenyl-7-ethyl-3,8,13,17-tetramethyl-17,18-dihydroporphyrin-21,23-diide-2-carboxylate | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-30 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | CCA-bound alpha-synuclein fibrils
To Be Published
|
|
7YQ8
 
 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | Authors: | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | Deposit date: | 2022-08-05 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
7YNZ
 
 | |
7YO0
 
 | | Cryo-EM structure of human Slo1-LRRC26 complex with Symmetry Expansion | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CALCIUM ION, Calcium-activated potassium channel subunit alpha-1, ... | | Authors: | Yamanouchi, D, Nureki, O. | | Deposit date: | 2022-08-01 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for dual allosteric gating modulation of Slo1-LRRC channel complex
To Be Published
|
|
7YO2
 
 | |
7YO5
 
 | |
5U0R
 
 | |
7YO1
 
 | |
7YO4
 
 | |
2IY5
 
 | | PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS complexed with tRNA and a phenylalanyl-adenylate analog | | Descriptor: | ADENOSINE-5'-[PHENYLALANINOL-PHOSPHATE], MAGNESIUM ION, PHENYLALANYL-TRNA SYNTHETASE ALPHA CHAIN, ... | | Authors: | Moor, N, Kotik-Kogan, O, Tworowski, D, Sukhanova, M, Safro, M. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of the ternary complex of phenylalanyl-tRNA synthetase with tRNAPhe and a phenylalanyl-adenylate analogue reveals a conformational switch of the CCA end.
Biochemistry, 45, 2006
|
|
3EWX
 
 | | K314A mutant of human orotidyl-5'-monophosphate decarboxylase in complex with 6-azido-UMP, degraded to BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine-5'-phosphate decarboxylase | | Authors: | Heinrich, D, Wittmann, J, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
3W7A
 
 | | Crystal Structure of azoreductase AzrC fin complex with sulfone-modified azo dye Acid Red 88 | | Descriptor: | 4-[(E)-(2-hydroxynaphthalen-1-yl)diazenyl]naphthalene-1-sulfonic acid, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yu, J, Ogata, D, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5U9D
 
 | | Discovery of a potent BTK inhibitor with a novel binding mode using parallel selections with a DNA-encoded chemical library | | Descriptor: | (R)-N-methyl-2-(3-((quinoxalin-6-ylamino)methyl)furan-2-carbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cuozzo, J.W, Centrella, P.A, Gikunju, D, Habeshian, S, Hupp, C.D, Keefe, A.D, Sigel, E, Soutter, H.H, Thomson, H.A, Zhang, Y, Clark, M.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library.
Chembiochem, 18, 2017
|
|
7ZGU
 
 | | Human NLRP3-deltaPYD hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Raisch, T, Machtens, D.A, Bresch, I.B, Eberhage, J, Prumbaum, D, Reubold, T.F, Raunser, S, Eschenburg, S. | | Deposit date: | 2022-04-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NEK7-independent NLRP3 inflammasome
To Be Published
|
|
5U9Q
 
 | | Ocellatin-LB1 | | Descriptor: | Ocellatin-LB1 | | Authors: | Gusmao, K.A.G, Santos, D.M, de Lima, M.E, Pilo-Veloso, D, Resende, J.M. | | Deposit date: | 2016-12-17 | | Release date: | 2017-12-13 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5U9V
 
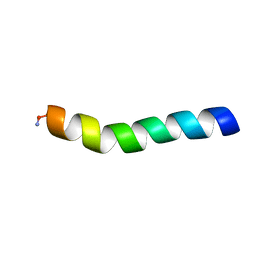 | | Ocellatin-LB1, solution structure in DPC micelle by NMR spectroscopy | | Descriptor: | Ocellatin-LB1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-29 | | Method: | SOLUTION NMR | | Cite: | Ocellatin-LB1
To Be Published
|
|
3FDQ
 
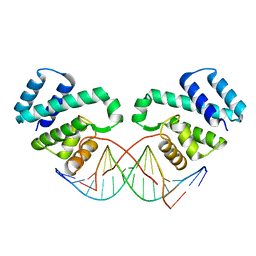 | |
3F5F
 
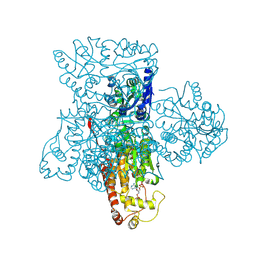 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7ZNX
 
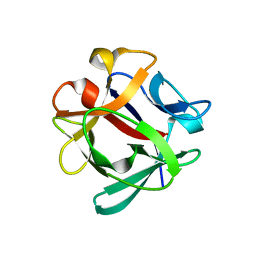 | |
