1YL8
 
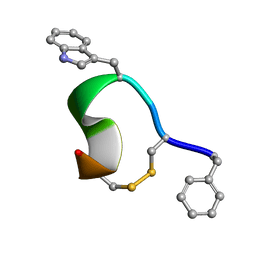 | | 3D Solution Structure of [Tyr3]Octreotate derivatives in DMSO | | Descriptor: | [Tyr3]Octreotate peptide | | Authors: | Spyroulias, G.A, Galanis, A.S, Petrou, C, Vahliotis, D, Sotiriou, P, Nikolopoulou, A, Nock, B, Maina, T, Cordopatis, P. | | Deposit date: | 2005-01-19 | | Release date: | 2005-09-20 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of [Tyr3]octreotate derivatives in DMSO: structure differentiation of peptide core due to chelate group attachment and biologically active conformation.
Med.Chem., 1, 2005
|
|
1YL9
 
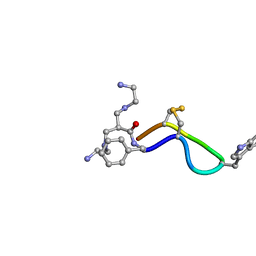 | | 3D Solution Structure of [Tyr3]Octreotate derivatives in DMSO | | Descriptor: | 3-[(2-AMINOETHYL)AMINO]-2-{[(2-AMINOETHYL)AMINO]METHYL}PROPANAL, [Tyr3]Octreotate | | Authors: | Spyroulias, G.A, Galanis, A.S, Petrou, C, Vahliotis, D, Sotiriou, P, Nikolopoulou, A, Nock, B, Maina, T, Cordopatis, P. | | Deposit date: | 2005-01-19 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of [Tyr3]octreotate derivatives in DMSO: structure differentiation of peptide core due to chelate group attachment and biologically active conformation.
Med.Chem., 1, 2005
|
|
1YMG
 
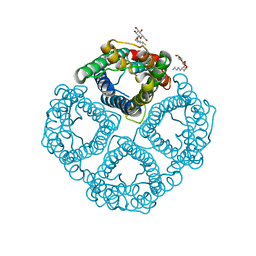 | | The Channel Architecture of Aquaporin O at 2.2 Angstrom Resolution | | Descriptor: | Lens fiber major intrinsic protein, nonyl beta-D-glucopyranoside | | Authors: | Harries, W.E.C, Akhavan, D, Miercke, L.J.W, Khademi, S, Stroud, R.M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Channel Architecture of Aquaporin 0 at a 2.2-A Resolution
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3ERD
 
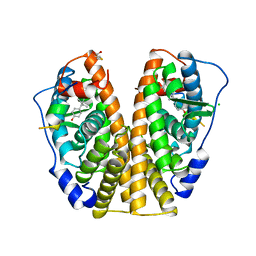 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH DIETHYLSTILBESTROL AND A GLUCOCORTICOID RECEPTOR INTERACTING PROTEIN 1 NR BOX II PEPTIDE | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIETHYLSTILBESTROL, ... | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
3E3M
 
 | | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi | | Descriptor: | Transcriptional regulator, LacI family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-07 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi
To be Published
|
|
3EUW
 
 | |
3E9G
 
 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
6NB9
 
 | | Amyloid-Beta (20-34) with L-isoaspartate 23 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Boyer, D.R, Zee, C.T, Richards, L.S, Cascio, D, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-08-07 | | Last modified: | 2022-09-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
6NIY
 
 | | A high-resolution cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | dal Maso, E, Glukhova, A, Zhu, Y, Garcia-Nafria, J, Tate, C.G, Atanasio, S, Reynolds, C.A, Ramirez-Aportela, E, Carazo, J.-M, Hick, C.A, Furness, S.G.B, Hay, D.L, Liang, Y.-L, Miller, L.J, Christopoulos, A, Wang, M.-W, Wootten, D, Sexton, P.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-01-23 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | The Molecular Control of Calcitonin Receptor Signaling.
Acs Pharmacol Transl Sci, 2, 2019
|
|
3EAR
 
 | |
3ES9
 
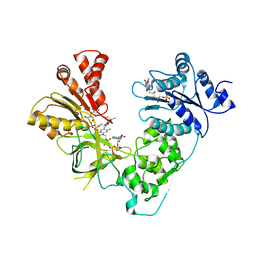 | | NADPH-Cytochrome P450 Reductase in an Open Conformation | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hamdane, D, Xia, C, Im, S.-C, Zhang, H, Kim, J.-J, Waskell, L. | | Deposit date: | 2008-10-05 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450
J.Biol.Chem., 284, 2009
|
|
3EUV
 
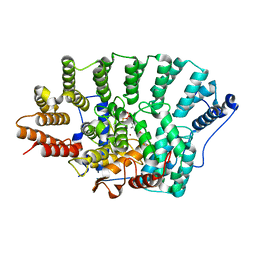 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10, W102T, Y154T) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
6NTS
 
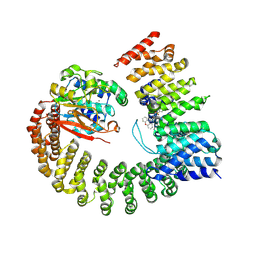 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
3EIP
 
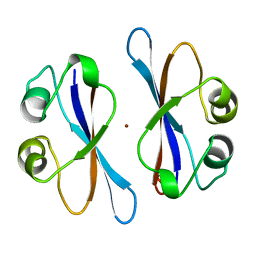 | | CRYSTAL STRUCTURE OF COLICIN E3 IMMUNITY PROTEIN: AN INHIBITOR TO A RIBOSOME-INACTIVATING RNASE | | Descriptor: | PROTEIN (COLICIN E3 IMMUNITY PROTEIN), ZINC ION | | Authors: | Li, C, Zhao, D, Djebli, A, Shoham, M. | | Deposit date: | 1999-03-29 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase.
Structure Fold.Des., 7, 1999
|
|
3EKD
 
 | |
3EM0
 
 | | Crystal structure of Zebrafish Ileal Bile Acid-Bindin Protein complexed with cholic acid (crystal form B). | | Descriptor: | CHOLIC ACID, Ileal Bile Acid-Binding Protein | | Authors: | Capaldi, S, Saccomani, G, Fessas, D, Signorelli, M, Perduca, M, Monaco, H.L. | | Deposit date: | 2008-09-23 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-Ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule.
J.Mol.Biol., 385, 2009
|
|
3EU5
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
6NJJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN14770 | | Descriptor: | (4-{[2-(3-chlorophenyl)-6-(trifluoromethyl)pyridin-4-yl]methyl}phenyl)acetic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
4CZV
 
 | | Structure of the Neurospora crassa Pan2 WD40 domain | | Descriptor: | CHLORIDE ION, GLYCEROL, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN2 | | Authors: | Peter, D, Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Asymmetric Pan3 Dimer Recruits a Single Pan2 Exonuclease to Mediate Mrna Deadenylation and Decay.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3E8M
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acylneuraminate cytidylyltransferase, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
3EAS
 
 | |
4CSV
 
 | | Tyrosine kinase AS - a common ancestor of Src and Abl bound to Gleevec | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, SRC-ABL TYROSINE KINASE ANCESTOR | | Authors: | Kutter, S, Wilson, C, Cruz, L, Agafonov, R.V, Hoemberger, M.S, Zorba, A, Kern, D. | | Deposit date: | 2014-03-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinase Dynamics. Using Ancient Protein Kinases to Unravel a Modern Cancer Drug'S Mechanism.
Science, 347, 2015
|
|
3EQM
 
 | |
3ERT
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH 4-HYDROXYTAMOXIFEN | | Descriptor: | 4-HYDROXYTAMOXIFEN, PROTEIN (ESTROGEN RECEPTOR ALPHA) | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-30 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
3EUL
 
 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
