3TEM
 
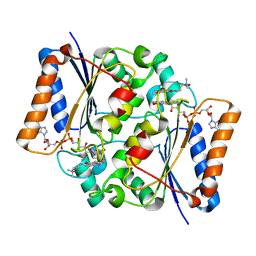 | | Quinone Oxidoreductase (NQ02) bound to the imidazoacridin-6-one 6a1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Dunstan, M.S, Leys, D. | | Deposit date: | 2011-08-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitors of NRH:Quinone Oxidoreductase 2 (NQO2): Crystal Structures, Biochemical Activity, and Intracellular Effects of Imidazoacridin-6-ones.
J.Med.Chem., 54, 2011
|
|
3D2M
 
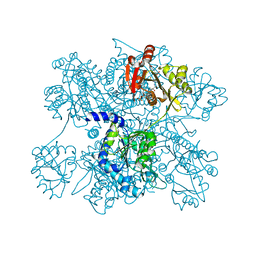 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-glutamate | | Descriptor: | COENZYME A, GLUTAMIC ACID, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
2GG1
 
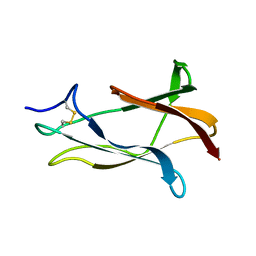 | | NMR solution structure of domain III of the E-protein of tick-borne Langat flavivirus (includes RDC restraints) | | Descriptor: | Genome polyprotein | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
5V5B
 
 | | KVQIINKKLD, Structure of the amyloid spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
5V5S
 
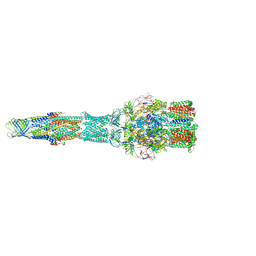 | | multi-drug efflux; membrane transport; RND superfamily; Drug resistance | | Descriptor: | Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, Outer membrane protein TolC | | Authors: | wang, Z, fan, G, Hryc, C.F, Blaza, J.N, Serysheva, I.I, Schmid, M.F, Chiu, W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-15 | | Release date: | 2017-04-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | An allosteric transport mechanism for the AcrAB-TolC Multidrug Efflux Pump.
Elife, 6, 2017
|
|
3CL9
 
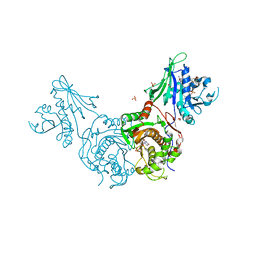 | | Structure of bifunctional TcDHFR-TS in complex with MTX | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS), ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3T62
 
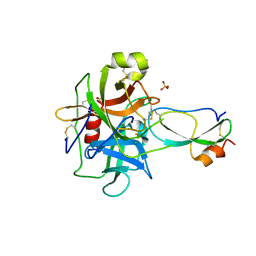 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
3T6P
 
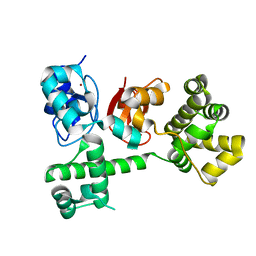 | | IAP antagonist-induced conformational change in cIAP1 promotes E3 ligase activation via dimerization | | Descriptor: | Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dueber, E.C, Schoeffler, A.J, Lingel, A, Elliott, M, Fedorova, A.V, Giannetti, A.M, Zobel, K, Maurer, B, Varfolomeev, E, Wu, P, Wallweber, H, Hymowitz, S, Deshayes, K, Vucic, D, Fairbrother, W.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination.
Science, 334, 2011
|
|
2FO8
 
 | | Solution structure of the Trypanosoma cruzi cysteine protease inhibitor chagasin | | Descriptor: | Chagasin | | Authors: | Salmon, D, do Aido-Machado, R, de Lima, A.A.P, Scharfstein, J, Oschkinat, H, Pires, J.R. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Trypanosoma cruzi Cysteine Protease Inhibitor Chagasin
J.Mol.Biol., 357, 2006
|
|
2FQH
 
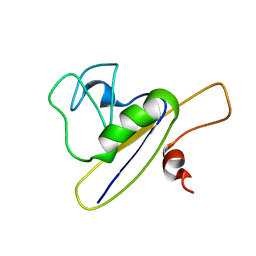 | | NMR structure of hypothetical protein TA0938 from Termoplasma acidophilum | | Descriptor: | Hypothetical protein TA0938 | | Authors: | Monleon, D, Esteve, V, Yee, A, Arrowsmith, C, Celda, B, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of hypothetical protein TA0938 from Thermoplasma acidophilum.
Proteins, 67, 2007
|
|
3CKI
 
 | | Crystal structure of the TACE-N-TIMP-3 complex | | Descriptor: | ADAM 17, Metalloproteinase inhibitor 3, SODIUM ION, ... | | Authors: | Wisniewska, M, Goettig, P, Maskos, K, Belouski, E, Winters, D, Hecht, R, Black, R, Bode, W. | | Deposit date: | 2008-03-15 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex.
J.Mol.Biol., 381, 2008
|
|
3V1Q
 
 | |
3V1U
 
 | | Crystal structure of a beta-ketoacyl reductase FabG4 from Mycobacterium tuberculosis H37Rv complexed with NAD+ and Hexanoyl-CoA at 2.5 Angstrom resolution | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, HEXANOYL-COENZYME A, ... | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
3UT9
 
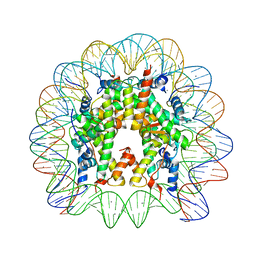 | | Crystal Structure of Nucleosome Core Particle Assembled with a Palindromic Widom '601' Derivative (NCP-601L) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3UWA
 
 | | Crystal structure of a probable peptide deformylase from synechococcus phage S-SSM7 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, RIIA-RIIB membrane-associated protein, ZINC ION | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
2GPT
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase in complex with tartrate and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase/ shikimate 5-dehydrogenase, L(+)-TARTARIC ACID, ... | | Authors: | Singh, S.A, Christendat, D. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic channeling in the shikimate pathway
Biochemistry, 45, 2006
|
|
3UWU
 
 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with glycerol-3-phosphate | | Descriptor: | CITRIC ACID, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
5ULC
 
 | | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328 | | Descriptor: | Bromodomain protein 1 | | Authors: | Wernimont, A.K, Amaya, M.F, Lam, A, Ali, A, Zhang, A.Z, Kenzina, L, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Hui, R, Walker, J.R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328
To be published
|
|
7ZHC
 
 | | Moss spermine/spermidine acetyl transferase (PpSSAT) in complex with AcetylCoA and polyethylen glycol | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, N-acetyltransferase domain-containing protein, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2022-04-06 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Biochemical and structural basis of polyamine, lysine and ornithine acetylation catalyzed by spermine/spermidine N-acetyl transferase in moss and maize.
Plant J., 114, 2023
|
|
3CQN
 
 | |
5UMU
 
 | | Crystal structure of the middle double PH domain of human FACT complex subunit SPT16 | | Descriptor: | ACETATE ION, FACT complex subunit SPT16, FORMIC ACID | | Authors: | Hu, Q, Thompson, J.R, Heroux, A, Su, D, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the middle double PH domain of human FACT complex subunit SPT16
To Be Published
|
|
3CPS
 
 | | Crystal structure of Cryptosporidium parvum glyceraldehyde-3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-01 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Cryptosporidium parvum glyceraldehyde-3-phosphate dehydrogenase.
To be Published
|
|
3S6G
 
 | | Crystal structures of Seleno-substituted mutant mmNAGS in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, MALONATE ION, ... | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6681 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
2GTH
 
 | | crystal structure of the wildtype MHV coronavirus non-structural protein nsp15 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
3ULC
 
 | |
