6E0V
 
 | | Apo crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriophage Phi92 gp150, CHLORIDE ION, ... | | Authors: | Leiman, P.G, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Plattner, M, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
3IW5
 
 | | Human p38 MAP Kinase in Complex with an Indole Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-(3-{[2-(2,3-dihydro-1,4-benzodioxin-6-ylamino)-2-oxoethyl]sulfanyl}-1H-indol-1-yl)ethyl]-3-(trifluoromethyl)benzamide, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
6DZZ
 
 | | Cryo-EM Structure of the wild-type human serotonin transporter in complex with ibogaine and 15B8 Fab in the inward conformation | | Descriptor: | (5beta)-12-methoxyibogamine, 15B8 antibody heavy chain, 15B8 antibody light chain, ... | | Authors: | Yang, D, Coleman, J.A, Gouaux, E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport.
Nature, 569, 2019
|
|
6E0W
 
 | | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-4,6-O-[(1R)-1-carboxyethylidene]-D-galactitol, Bacteriophage Phi92 gp150, ... | | Authors: | Plattner, M, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Leiman, P.G, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
3ISR
 
 | | The Crystal Structure of a Putative Cysteine Protease from Cytophaga hutchinsonii to 1.9A | | Descriptor: | Transglutaminase-like enzymes, putative cysteine protease | | Authors: | Stein, A.J, Bigelow, L, Trevino, D, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of a Putative Cysteine Protease from Cytophaga hutchinsonii to 1.9A
To be Published
|
|
3ITA
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in acyl-enzyme complex with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase dacC, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3IDU
 
 | | Crystal Structure of the CARDB domain of the PF1109 protein in complex with di-metal ions from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR193A | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target PfR193A
To be Published
|
|
3I9I
 
 | | Crystal structure of human transthyretin variant A25T - #2 | | Descriptor: | TRIETHYLENE GLYCOL, Transthyretin | | Authors: | Palmieri, L.C, Freire, J.B.B, Palhano, F.L, Azevedo, E.P.C, Foguel, D, Lima, L.M.T.R. | | Deposit date: | 2009-07-11 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human transthyretin variant A25T - #2
To be Published
|
|
6RRH
 
 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6ELP
 
 | |
3IAE
 
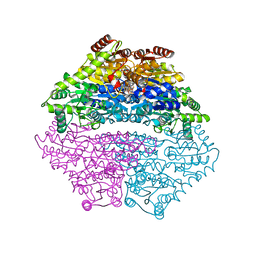 | | Structure of benzaldehyde lyase A28S mutant with benzoylphosphonate | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzaldehyde lyase, CALCIUM ION | | Authors: | Brandt, G.S, Petsko, G.A, Ringe, D, McLeish, M.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site engineering of benzaldehyde lyase shows that a point mutation can confer both new reactivity and susceptibility to mechanism-based inhibition.
J.Am.Chem.Soc., 132, 2010
|
|
6ELX
 
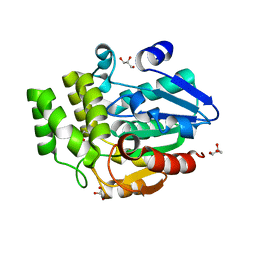 | | Oryza sativa DWARF14 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Andersson, I, Carlsson, G.H, Hasse, D. | | Deposit date: | 2017-09-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The elusive ligand complexes of the DWARF14 strigolactone receptor.
J. Exp. Bot., 69, 2018
|
|
6EQU
 
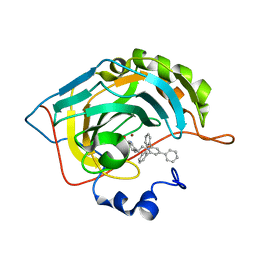 | | X-Ray crystal structure of the human carbonic anhydrase II adduct with a membrane-impermeant inhibitor | | Descriptor: | 4-[2-(2,4,6-triphenylpyridin-1-ium-1-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human carbonic anhydrase II adduct with 1-(4-sulfamoylphenyl-ethyl)-2,4,6-triphenylpyridinium perchlorate, a membrane-impermeant, isoform selective inhibitor.
J Enzyme Inhib Med Chem, 33, 2018
|
|
3IIR
 
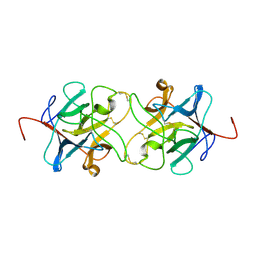 | | Crystal Structure of Miraculin like protein from seeds of Murraya koenigii | | Descriptor: | Trypsin inhibitor | | Authors: | Gahloth, D, Selvakumar, P, Shee, C, Kumar, P, Sharma, A.K. | | Deposit date: | 2009-08-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cloning, sequence analysis and crystal structure determination of a miraculin-like protein from Murraya koenigii
Arch.Biochem.Biophys., 494, 2010
|
|
3IK8
 
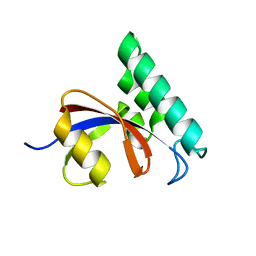 | |
6E59
 
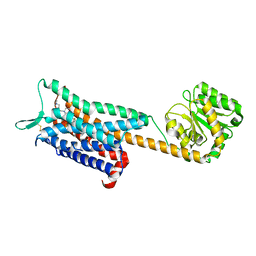 | | Crystal structure of the human NK1 tachykinin receptor | | Descriptor: | 1-(4-{[(2R,3S)-2-{(1R)-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)morpholin-4-yl]methyl}-1H-1,2,3-triazol-5-yl)-N,N-dimethylmethanamine, Substance-P receptor, GlgA glycogen synthase, ... | | Authors: | Yin, J, Clark, L, Chapman, K, Shao, Z, Borek, D, Xu, Q, Wang, J, Rosenbaum, D.M. | | Deposit date: | 2018-07-19 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human NK1tachykinin receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3IF6
 
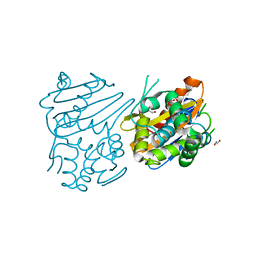 | | Crystal structure of OXA-46 beta-lactamase from P. aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, L(+)-TARTARIC ACID, ... | | Authors: | Docquier, J.D, Benvenuti, M, Calderone, V, Giuliani, F, Kapetis, D, De Luca, F, Rossolini, G.M, Mangani, S. | | Deposit date: | 2009-07-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the narrow-spectrum OXA-46 class D beta-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates
Antimicrob.Agents Chemother., 54, 2010
|
|
6RKA
 
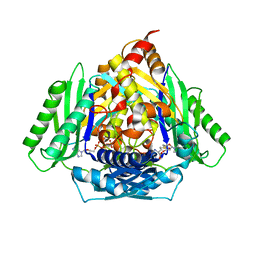 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Methionine adenosyltransferase, PHOSPHATE ION, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
3IPI
 
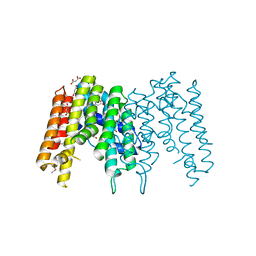 | | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei | | Descriptor: | Geranyltranstransferase, MALONIC ACID | | Authors: | Kumaran, D, Mohammed, M.B, Brown, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-08 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Geranyltranstransferase from the Methanosarcina mazei
To be Published
|
|
6E9Z
 
 | | DHF119 filament | | Descriptor: | DHF119 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6RPK
 
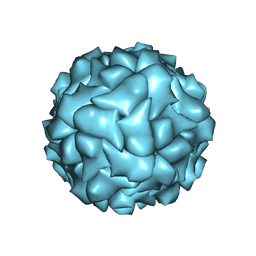 | |
6EBA
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Unoccupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-08-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6RPO
 
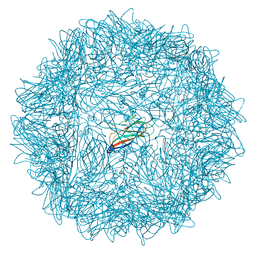 | |
6RPS
 
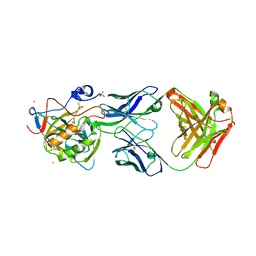 | | X-ray crystal structure of carbonic anhydrase XII complexed with a theranostic monoclonal antibody fragment | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Alterio, V, Esposito, D, De Simone, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Biochemical and Structural Insights into Carbonic Anhydrase XII/Fab6A10 Complex.
J.Mol.Biol., 431, 2019
|
|
6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | Descriptor: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
