1M0J
 
 | | solution structure of the beta domain of mt_nc | | Descriptor: | CADMIUM ION, metallothionein MT_nc | | Authors: | Capasso, C, Carginale, V, Crescenzi, O, Di Maro, D, Parisi, E, Spadaccini, R, Temussi, P.A. | | Deposit date: | 2002-06-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MT_nc, a Novel Metallothionein from the Antarctic Fish Notothenia coriiceps.
Structure, 11, 2003
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
6L1E
 
 | |
1M54
 
 | | CYSTATHIONINE-BETA SYNTHASE: REDUCED VICINAL THIOLS | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Taoka, S, Lepore, B.W, Kabil, O, Ojha, S, Ringe, D, Banerjee, R. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HUMAN CYSTATHIONINE BETA-SYNTHASE IS A HEME SENSOR PROTEIN. EVIDENCE THAT THE
REDOX SENSOR IS HEME AND NOT THE VICINAL CYSTEINES IN THE CXXC MOTIF SEEN IN THE CRYSTAL STRUCTURE OF THE TRUNCATED ENZYME
BIOCHEMISTRY, 41, 2002
|
|
6LA8
 
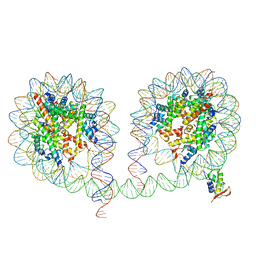 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
1MOJ
 
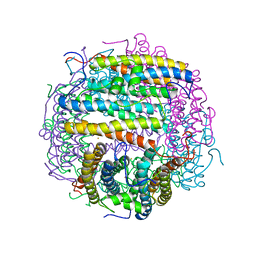 | | Crystal structure of an archaeal dps-homologue from Halobacterium salinarum | | Descriptor: | Dps-like ferritin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2002-09-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: Structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1IIZ
 
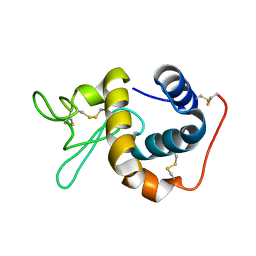 | | Crystal Structure of the Induced Antibacterial Protein from Tasar Silkworm, Antheraea mylitta | | Descriptor: | LYSOZYME | | Authors: | Jain, D, Nair, D.T, Swaminathan, G.J, Abraham, E.G, Nagaraju, J, Salunke, D.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the induced antibacterial protein from tasar silkworm, Antheraea mylitta. Implications to molecular evolution.
J.Biol.Chem., 276, 2001
|
|
3UID
 
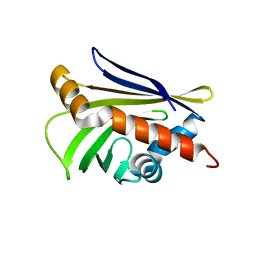 | | Crystal Structure of Protein Ms6760 from Mycobacterium smegmatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Bajaj, R.A, Miallau, L, Cascio, D, Arbing, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-11-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Crystal structure of the toxin Msmeg_6760, the structural homolog of Mycobacterium tuberculosis Rv2035, a novel type II toxin involved in the hypoxic response.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
1IY1
 
 | | Crystal structure of the FtsH ATPase domain with ADP from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent metalloprotease FtsH | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IE8
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IEH
 
 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
3UMI
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with zinc | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
1MI1
 
 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | Descriptor: | Neurobeachin | | Authors: | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
3UMH
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with cadmium | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
1M1P
 
 | | P21 crystal structure of the tetraheme cytochrome c3 from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, Small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
1M52
 
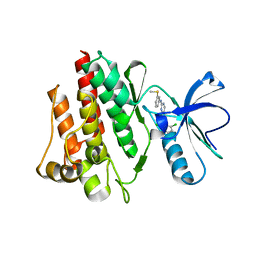 | | Crystal Structure of the c-Abl Kinase domain in complex with PD173955 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2,6-DICHLORO-PHENYL)-8-METHYL-2-(3-METHYLSULFANYL-PHENYLAMINO)-8H-PYRIDO[2,3-D]PYRIMIDIN-7-ONE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Nagar, B, Bornmann, W, Pellicena, P, Schindler, T, Veach, D, Miller, W.T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2002-07-08 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Kinase Domain of c-Abl in Complex with the Small Molecule Inhibitors PD173955 and Imatinib (STI-571)
Cancer Res., 62, 2002
|
|
1M7B
 
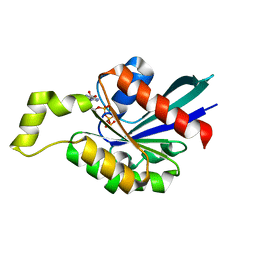 | | Crystal structure of Rnd3/RhoE: functional implications | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rnd3/RhoE small GTP-binding protein | | Authors: | Fiegen, D, Blumenstein, L, Stege, P, Vetter, I.R, Ahmadian, M.R. | | Deposit date: | 2002-07-19 | | Release date: | 2002-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Rnd3/RhoE: functional implications
FEBS LETT., 525, 2002
|
|
1M5H
 
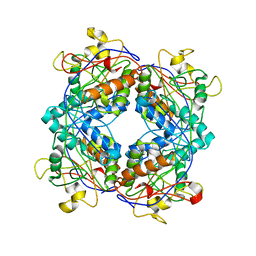 | | Formylmethanofuran:tetrahydromethanopterin formyltransferase from Archaeoglobus fulgidus | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase, POTASSIUM ION | | Authors: | Mamat, B, Roth, A, Grimm, C, Ermler, U, Tziatzios, C, Schubert, D, Thauer, R.K, Shima, S. | | Deposit date: | 2002-07-09 | | Release date: | 2002-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
Protein Sci., 11, 2002
|
|
1M9W
 
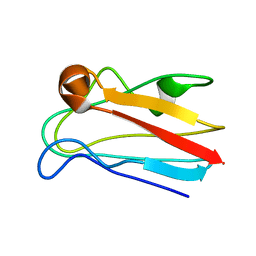 | |
1MBV
 
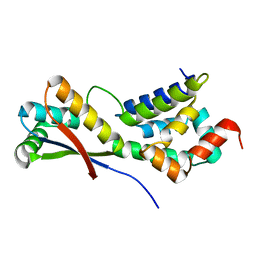 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN HETERODIMER TETRAGONAL FORM | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, Protein yljA | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of AAA+ Chaperone ClpA
J.Biol.Chem., 277, 2002
|
|
1MJD
 
 | | Structure of N-terminal domain of human doublecortin | | Descriptor: | DOUBLECORTIN | | Authors: | Kim, M.H, Cierpicki, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z.S. | | Deposit date: | 2002-08-27 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
1MFW
 
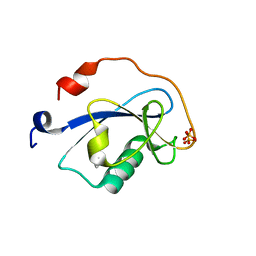 | | STRUCTURE OF N-TERMINAL DOUBLECORTIN DOMAIN FROM DCLK: SELENOMETHIONINE LABELED PROTEIN | | Descriptor: | DOUBLECORTIN-LIKE KINASE (N-TERMINAL DOMAIN), SULFATE ION | | Authors: | Kim, M.H, Cierpickil, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z. | | Deposit date: | 2002-08-13 | | Release date: | 2003-04-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
1MG2
 
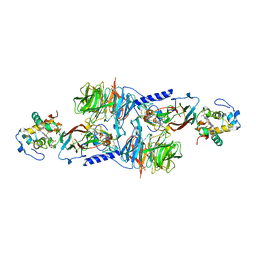 | | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN | | Descriptor: | Amicyanin, COPPER (II) ION, CYTOCHROME C-L, ... | | Authors: | Sun, D, Chen, Z.W, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-08-14 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | MUTATION OF AlPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN
Biochemistry, 41, 2002
|
|
4MC1
 
 | | HIV protease in complex with SA526P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4S)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
1MN9
 
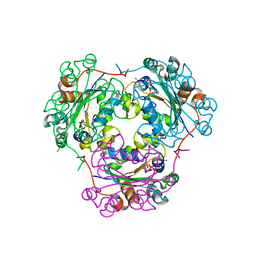 | | NDP kinase mutant (H122G) complex with RTP | | Descriptor: | MAGNESIUM ION, NDP kinase, RIBAVIRIN TRIPHOSPHATE | | Authors: | Gallois-montbrun, S, Chen, Y, Dutartre, H, Morera, S, Guerreiro, C, Mulard, L, Schneider, B, Janin, J, Canard, B, Veron, M, Deville-bonne, D. | | Deposit date: | 2002-09-05 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis of the Activation of Ribavirin Analogs by NDP Kinase: Comparison with Other Ribavirin Targets
MOL.PHARMACOL., 63, 2003
|
|
