4HHU
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR280. | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, OR280, TETRAETHYLENE GLYCOL | | Authors: | Vorobiev, S, Lew, S, Lin, Y.-R, Seetharaman, J, Castelllanos, J, Maglaqui, M, Xiao, R, Lee, D, Koga, N, Koga, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-10 | | Release date: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Engineered Protein OR280.
To be Published
|
|
2CIR
 
 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, HEXOSE-6-PHOSPHATE MUTAROTASE | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
2CIC
 
 | | THE CRYSTAL STRUCTURE OF A COMPLEX OF CAMPYLOBACTER JEJUNI DUTPASE WITH SUBSTRATE ANALOGUE DUPNHPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2006-03-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
2CI6
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I bound with Zinc low pH | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, ZINC ION | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inbitors
Structure, 14, 2006
|
|
2CHF
 
 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND THE MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
2CHM
 
 | | Crystal structure of N2 substituted pyrazolo pyrimidinones - a flipped binding mode in PDE5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2-(BUT-3-EN-1-YLOXY)-5-(1-HYDROXYVINYL)PYRIDIN-3-YL]-3-ETHYL-2-(1-ETHYLAZETIDIN-3-YL)-1,2,6,7A-TETRAHYDRO-7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, CGMP-SPECIFIC 3', ... | | Authors: | Allerton, C.M.N, Barber, C.G, Beaumont, K.C, Brown, D.G, Cole, S.M, Ellis, D, Lane, C.A.L, Maw, G.N, Mount, N.M, Rawson, D.J, Robinson, C.M, Street, S.D.A, Summerhill, N.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Series of Potent and Selective Pde5 Inhibitors with Potential for High and Dose-Independent Oral Bioavailability
J.Med.Chem., 49, 2006
|
|
2CLE
 
 | | Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2- amino-1-ethylphosphate (F6) - lowF6 complex | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
4HQD
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR265. | | Descriptor: | Engineered Protein OR265 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal Structure of Engineered Protein OR265.
To be Published
|
|
2C7O
 
 | |
1JRZ
 
 | | Crystal structure of Arg402Tyr mutant flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C, FUMARIC ACID, ... | | Authors: | Mowat, C.G, Moysey, R, Miles, C.S, Leys, D, Doherty, M.K, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2001-08-15 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase.
Biochemistry, 40, 2001
|
|
2CLK
 
 | | Tryptophan Synthase in complex with D-glyceraldehyde 3-phosphate (G3P) | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
6LYD
 
 | | Crystal Structure of mimivirus UNG Y322L in complex with UGI | | Descriptor: | Probable uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Selective interactions between mimivirus uracil-DNA glycosylase and inhibitory proteins determined by a single amino acid.
J.Struct.Biol., 211, 2020
|
|
6M37
 
 | |
2CLS
 
 | | The crystal structure of the human RND1 GTPase in the active GTP bound state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RHO-RELATED GTP-BINDING PROTEIN RHO6 | | Authors: | Pike, A.C.W, Yang, X, Colebrook, S, Gileadi, O, Sobott, F, Bray, J, Wen Hwa, L, Marsden, B, Zhao, Y, Schoch, G, Elkins, J, Debreczeni, J.E, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of the Human Rnd1 Gtpase in the Active GTP Bound State
To be Published
|
|
2CC2
 
 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 5'deoxyadenosine | | Descriptor: | 5'-DEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, CHLORIDE ION | | Authors: | Mcewan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.R, O'Hagan, D, Spencer, J, Naismith, J.H. | | Deposit date: | 2006-01-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
5LNY
 
 | | HSP90 WITH indazole derivative | | Descriptor: | 6-Hydroxy-3-(piperidine-1-carbonyl)-1H-indazole-5-carboxylic acid methyl-(4-morpholin-4-yl-phenyl)-amide, Heat shock protein HSP 90-alpha | | Authors: | Graedler, U, Amaral, M, Schuetz, D. | | Deposit date: | 2016-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6AGK
 
 | | The structure of CH-II-77-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, H, Arnst, K, Wang, Y, Miller, D, Li, W. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
2DVH
 
 | | THE Y64A MUTANT OF CYTOCHROME C553 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH, NMR, 39 STRUCTURES | | Descriptor: | CYTOCHROME C-553, HEME C | | Authors: | Sebban-Kreuzer, C, Blackledge, M.J, Dolla, A, Marion, D, Guerlesquin, F. | | Deposit date: | 1998-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of ferrocytochrome c553 from Desulfovibrio vulgaris studied by NMR spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 245, 1995
|
|
2DEA
 
 | |
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
1JRY
 
 | | Crystal structure of Arg402Lys mutant flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C, FUMARIC ACID, ... | | Authors: | Mowat, C.G, Moysey, R, Miles, C.S, Leys, D, Doherty, M.K, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2001-08-15 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase.
Biochemistry, 40, 2001
|
|
2DKD
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
6LYE
 
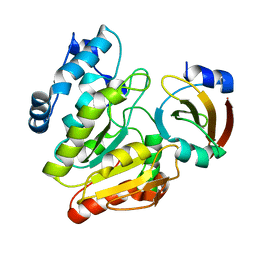 | | Crystal Structure of mimivirus UNG Y322F in complex with UGI | | Descriptor: | Probable uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Selective interactions between mimivirus uracil-DNA glycosylase and inhibitory proteins determined by a single amino acid.
J.Struct.Biol., 211, 2020
|
|
6LTN
 
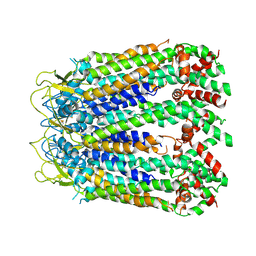 | | cryo-EM structure of C-terminal truncated human Pannexin1 | | Descriptor: | Pannexin-1 | | Authors: | Mou, L.Q, Ke, M, Xiao, Q.J, Wu, J.P, Deng, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for gating mechanism of Pannexin 1 channel.
Cell Res., 30, 2020
|
|
6M3L
 
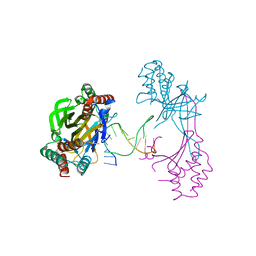 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(nonspecific) complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*CP*GP*AP*TP*TP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*GP*CP*G)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
