3CTA
 
 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
3CS6
 
 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | Authors: | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, P, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
3V1Z
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP*G)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | Descriptor: | EHEE_rd1_0284 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
2GSV
 
 | | X-Ray Crystal Structure of Protein YvfG from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR478. | | Descriptor: | Hypothetical protein yvfG, SULFATE ION | | Authors: | Forouhar, F, Su, M, Jayaraman, S, Wang, D, Fang, Y, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Hypothetical Protein YvfG from Bacillus subtilis, Northeast Structural Genomics Target SR478
To be Published
|
|
2GU1
 
 | | Crystal structure of a zinc containing peptidase from vibrio cholerae | | Descriptor: | SODIUM ION, ZINC ION, Zinc peptidase | | Authors: | Sugadev, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative lysostaphin peptidase from Vibrio cholerae.
Proteins, 72, 2008
|
|
3D04
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with sakuranetin | | Descriptor: | (2S)-5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-2,3-dihydro-4H-chromen-4-one, (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, ... | | Authors: | Zhang, L, Kong, Y, Wu, D, Shen, X, Jiang, H. | | Deposit date: | 2008-05-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
3V6P
 
 | | Crystal structure of the DNA-binding domain of dHax3, a TAL effector | | Descriptor: | dHax3 | | Authors: | Deng, D, Yan, C.Y, Pan, X.J, Wang, J.W, Shi, Y.G, Yan, N. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Sequence-Specific Recognition of DNA by TAL Effectors
Science, 2012
|
|
7Z0W
 
 | | E. coli NfsA bound to NADP+ | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
3V71
 
 | | Crystal structure of PUF-6 in complex with 5BE13 RNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Puf (Pumilio/fbf) domain-containing protein 7, confirmed by transcript evidence, ... | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
2KBE
 
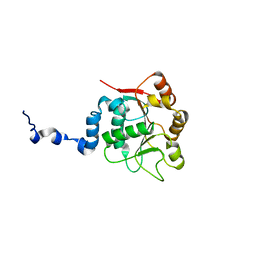 | |
3D2P
 
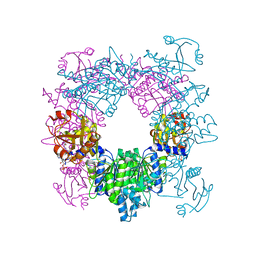 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-arginine | | Descriptor: | ARGININE, COENZYME A, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
3S6H
 
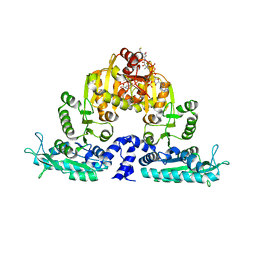 | | Crystal structure of native mmNAGS/k | | Descriptor: | COENZYME A, GLUTAMIC ACID, N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3UTA
 
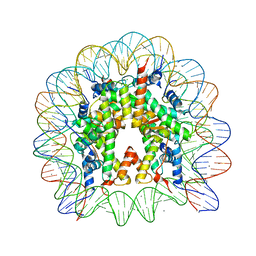 | | Crystal Structure of Nucleosome Core Particle Assembled with an Alpha-Satellite Sequence Containing Two TTAAA elements (NCP-TA2) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3DLP
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, Mutant D402P, bound to 4CB | | Descriptor: | 4-CHLORO-BENZOIC ACID, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Cao, J, Reger, A.S, Lu, X, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2008-06-28 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase
Biochemistry, 48, 2009
|
|
5V5N
 
 | | Crystal structure of Takinib bound to TAK1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N~1~-(1-propyl-1,3-dihydro-2H-benzimidazol-2-ylidene)benzene-1,3-dicarboxamide | | Authors: | Gurbani, D, Westover, K, Bera, A.K. | | Deposit date: | 2017-03-14 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Takinib, a Selective TAK1 Inhibitor, Broadens the Therapeutic Efficacy of TNF-alpha Inhibition for Cancer and Autoimmune Disease.
Cell Chem Biol, 24, 2017
|
|
2KL8
 
 | | Solution NMR Structure of de novo designed ferredoxin-like fold protein, Northeast Structural Genomics Consortium Target OR15 | | Descriptor: | OR15 | | Authors: | Liu, G, Koga, N, Jiang, M, Koga, R, Xiao, R, Ciccosanti, C, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
5V7G
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Mason, D.V, Handing, K.B, Kutner, J, Matelska, D, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V8E
 
 | | Structure of Bacillus cereus PatB1 | | Descriptor: | Bacillus cereus PatB1, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
3UW6
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR120 | | Descriptor: | Alanine racemase | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-08 | | Last modified: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
3SBA
 
 | | Zn-mediated Hexamer of T4 Lysozyme R76H/R80H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, Lysozyme, ZINC ION | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
7Z0L
 
 | | IL-27 signalling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Jin, Y, Gardner, S, Bubeck, D. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-27 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the assembly and activation of the IL-27 signaling complex.
Embo Rep., 23, 2022
|
|
2K69
 
 | |
3DQ0
 
 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-(3-methoxy-phenyl)adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and biological activity of novel purine-derived inhibitor of cytokinin oxidase/dehydrogenase and its potential use for in vivo studies
To be Published
|
|
7ZCS
 
 | | Nitrite-bound MSOX movie series dataset 65 (52 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - water ligand (final) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
