4V6T
 
 | | Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Yamamoto, H, Rother, K, Wittek, D, Pech, M, Mielke, T, Loerke, J, Scheerer, P, Ivanov, P, Teraoka, Y, Shpanchenko, O, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The complex of tmRNA-SmpB and EF-G on translocating ribosomes.
Nature, 485, 2012
|
|
4V1N
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4V4Z
 
 | | 70S Thermus thermophilous ribosome functional complex with mRNA and E- and P-site tRNAs at 4.5A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4V5D
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
5MW6
 
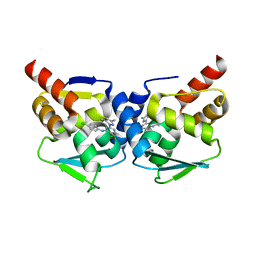 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
7A2B
 
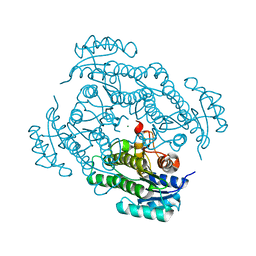 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q207D | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Bischoff, D, Hermann, J, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-08-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6HKN
 
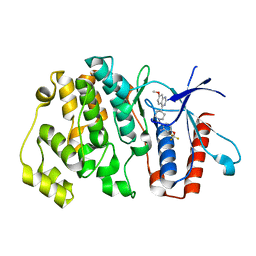 | | Crystal structure of Compound 35 with ERK5 | | Descriptor: | Mitogen-activated protein kinase 7, [2-azanyl-4-(trifluoromethyloxy)phenyl]-[4-(7-methoxyquinazolin-4-yl)piperidin-1-yl]methanone | | Authors: | Nguyen, D, Lemos, C, Wortmann, L, Eis, K, Holton, S.J, Boemer, U, Lechner, C, Prechtl, S, Suelze, D, Siegel, F, Prinz, F, Lesche, R, Nicke, B, Mumberg, D, Bauser, M, Haegebarth, A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine Based in Vitro Probe BAY-885 for the Kinase ERK5.
J. Med. Chem., 62, 2019
|
|
5FI8
 
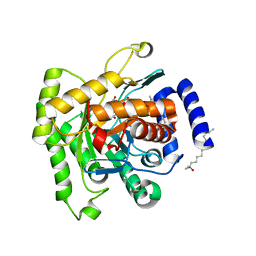 | | Crystal structure of plasmodium falciparum dihydroorotate dehydrogenase bounded with DSM422 (Tetrahydro-2-naphthyl and 2-indanyl triazolopyrimidine) | | Descriptor: | 2-[1,1-bis(fluoranyl)ethyl]-~{N}-[(2~{S})-7-bromanyl-1,2,3,4-tetrahydronaphthalen-2-yl]-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Kokkonda, S, Tomchick, D, Phillips, M. | | Deposit date: | 2015-12-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Tetrahydro-2-naphthyl and 2-Indanyl Triazolopyrimidines Targeting Plasmodium falciparum Dihydroorotate Dehydrogenase Display Potent and Selective Antimalarial Activity.
J.Med.Chem., 59, 2016
|
|
6PUW
 
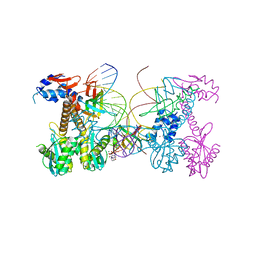 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and Bictegravir (BIC) | | Descriptor: | Bictegravir, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUZ
 
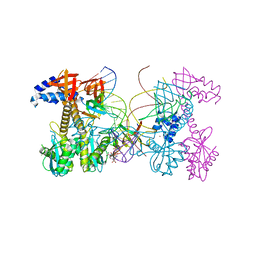 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUY
 
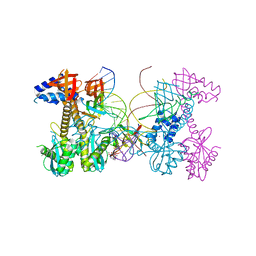 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUT
 
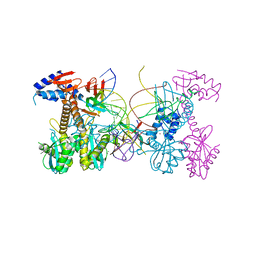 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with calcium | | Descriptor: | CALCIUM ION, Chimeric Sso7d and HIV-1 integrase, ZINC ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
3BWJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor lead compound Arc-1034 | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-N-(6-{[(1R)-4-carbamimidamido-1-{[(1R)-4-carbamimidamido-1-carbamoylbutyl]carbamoyl}butyl]amino}-6-oxohexyl)-3,4-dihydroxytetrahydrofuran-2-carboxamide, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Lavogina, D, Koenig, N, Uri, A, Bossemeyer, D. | | Deposit date: | 2008-01-09 | | Release date: | 2009-02-03 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases.
J.Med.Chem., 52, 2009
|
|
6HKM
 
 | | Crystal structure of Compound 1 with ERK5 | | Descriptor: | Mitogen-activated protein kinase 7, [4-(6,7-dimethoxyquinazolin-4-yl)piperidin-1-yl]-[4-(trifluoromethyloxy)phenyl]methanone | | Authors: | Nguyen, D, Lemos, C, Wortmann, L, Eis, K, Holton, S.J, Boemer, U, Lechner, C, Prechtl, S, Suelze, D, Siegel, F, Prinz, F, Lesche, R, Nicke, B, Mumberg, D, Bauser, M, Haegebarth, A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine Based in Vitro Probe BAY-885 for the Kinase ERK5.
J. Med. Chem., 62, 2019
|
|
5WYH
 
 | | Crystal structure of RidL(1-200) complexed with VPS29 | | Descriptor: | GLYCEROL, Interaptin, Vacuolar protein sorting-associated protein 29 | | Authors: | Yao, J, Sun, Q, Jia, D. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanism of inhibition of retromer transport by the bacterial effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8OFB
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch, hexagonal form | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Reverse gyrase, ... | | Authors: | Klostermeier, D, Rasche, R, Mhaindarkar, V, Kummel, D, Rudolph, M.G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1CP7
 
 | | AMINOPEPTIDASE FROM STREPTOMYCES GRISEUS | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, ZINC ION | | Authors: | Gilboa, R, Greenblatt, H.M, Perach, M, Spungin-Bialik, A, Lessel, U, Schomburg, D, Blumberg, S, Shoham, G. | | Deposit date: | 1999-06-10 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Interactions of Streptomyces griseus aminopeptidase with a methionine product analogue: a structural study at 1.53 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1D4K
 
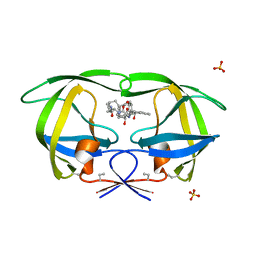 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1CSB
 
 | |
5DRB
 
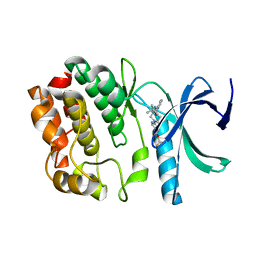 | | Crystal structure of WNK1 in complex with WNK463 | | Descriptor: | N-tert-butyl-1-(1-{5-[5-(trifluoromethyl)-1,3,4-oxadiazol-2-yl]pyridin-2-yl}piperidin-4-yl)-1H-imidazole-5-carboxamide, Serine/threonine-protein kinase WNK1 | | Authors: | Kohls, D, Xie, X. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Small-molecule WNK inhibition regulates cardiovascular and renal function.
Nat.Chem.Biol., 12, 2016
|
|
1A0G
 
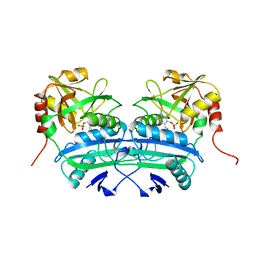 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAMINE-5'-PHOSPHATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, D-AMINO ACID AMINOTRANSFERASE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
8TBH
 
 | | Tricomplex of RMC-7977, KRAS G12R, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Bar Ziv, T, Zhang, D, Tomlinson, A.C.A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
