8OL5
 
 | | Murine type II Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL7
 
 | | MurineArc type I Abeta fibril from tg-APPArcSwe mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLG
 
 | | DI2 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLO
 
 | | Murine type III Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
2RLQ
 
 | | NMR structure of CCP modules 2-3 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-29 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
1TH3
 
 | | Crystal structure of NADPH depleted bovine live catalase complexed with cyanide | | Descriptor: | CYANIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Balasundaresan, D, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
7UHB
 
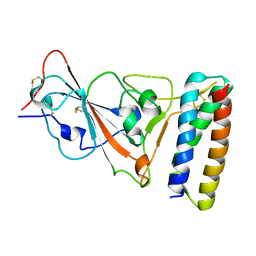 | |
5CX9
 
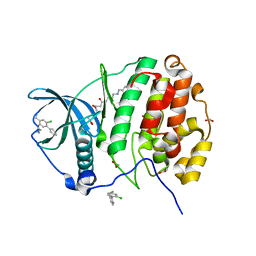 | | Crystal structure of CK2alpha with (methyl 4-((3-(3-chloro-4-(phenyl)benzylamino)propyl)amino)-4-oxobutanoate bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5LAV
 
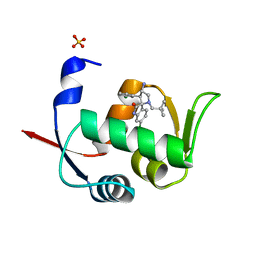 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) in complex with compound 6b | | Descriptor: | (3~{S},3'~{S},4'~{S})-4'-azanyl-6-chloranyl-3'-(3-chlorophenyl)-1'-(2,2-dimethylpropyl)spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAY
 
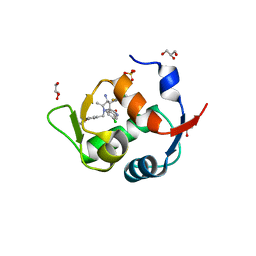 | | Discovery of New Natural-product-inspired Spiro-oxindole Compounds as Orally Active Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 6g | | Descriptor: | (3~{S},3'~{S},4'~{S},5'~{S})-4'-azanyl-6-chloranyl-3'-(3-chloranyl-2-fluoranyl-phenyl)-1'-[(3-ethoxyphenyl)methyl]-5'-methyl-spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
1AHS
 
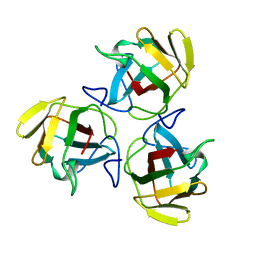 | |
1WOG
 
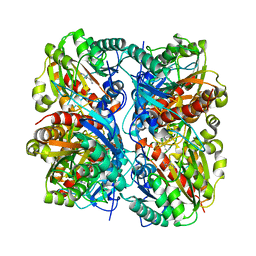 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | HEXANE-1,6-DIAMINE, MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1AI8
 
 | | HUMAN ALPHA-THROMBIN TERNARY COMPLEX WITH THE EXOSITE INHIBITOR HIRUGEN AND ACTIVE SITE INHIBITOR PHCH2OCO-D-DPA-PRO-BOROMPG | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN IIIB, ... | | Authors: | Skordalakes, E, Dodson, G, Elgendy, S, Goodwin, C.A, Green, D, Tyrrel, R, Scully, M.F, Freyssinet, J, Kakkar, V.V, Deadman, J. | | Deposit date: | 1997-05-01 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships.
Protein Sci., 1, 1992
|
|
5DEQ
 
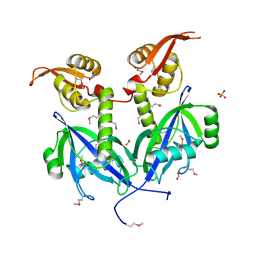 | | Crystal structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with L-arabinose | | Descriptor: | FORMIC ACID, SULFATE ION, TRANSCRIPTIONAL REGULATOR AraR, ... | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5XSG
 
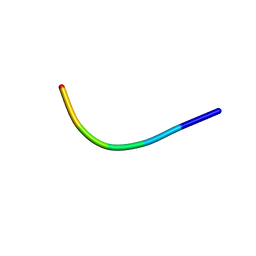 | | Ultrahigh resolution structure of FUS (37-42) SYSGYS determined by MicroED | | Descriptor: | RNA-binding protein FUS | | Authors: | Luo, F, Gui, X, Zhou, H, Li, D, Li, X, Liu, C. | | Deposit date: | 2017-06-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.73 Å) | | Cite: | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1ALY
 
 | |
7ZFE
 
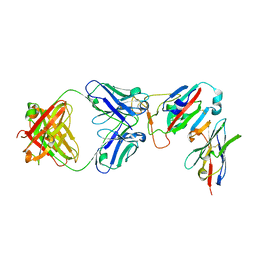 | | SARS-CoV-2 Omicron RBD in complex with Omi-32 Fab and nanobody C1 | | Descriptor: | Nanobody C1, Omi-32 heavy chain, Omi-32 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFD
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-25 Fab | | Descriptor: | Omi-25 heavy chain, Omi-25 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFF
 
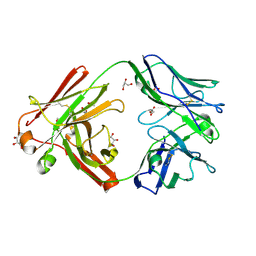 | | Omi-42 Fab | | Descriptor: | GLYCEROL, Omi-42 Heavy chain, Omi-42 light chain | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFC
 
 | | SARS-CoV-2 Beta RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | Nanobody C1, Omi-18 heavy chain, Omi-18 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF6
 
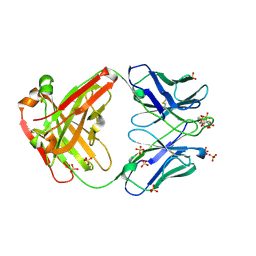 | | Omi-12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
5DDH
 
 | |
7ZF9
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab (P21) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 heavy chain, COVOX-150 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF8
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF5
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-12 and Beta-54 Fabs | | Descriptor: | Beta-54 heavy chain, Beta-54 light chain, Omi-12 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (5.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
