5AC7
 
 | | S. enterica HisA with mutations D7N, D10G, dup13-15 | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SODIUM ION, ... | | 著者 | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | 登録日 | 2015-08-12 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5AB3
 
 | | S.enterica HisA mutant D7N, D10G, dup13-15, Q24L, G102A | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SODIUM ION, [(2R,3S,4R,5R)-5-[4-AMINOCARBONYL-5-[[(Z)-[(3R,4R)-3,4-DIHYDROXY-2-OXO-5-PHOSPHONOOXY-PENTYL]IMINOMETHYL]AMINO]IMIDAZOL-1-YL]-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | 著者 | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | 登録日 | 2015-07-31 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6Z2M
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 antibody, Spike glycoprotein, ... | | 著者 | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | 登録日 | 2020-05-17 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published
|
|
6YZ7
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | 著者 | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | 登録日 | 2020-05-06 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
4YYZ
 
 | | 11B-HYDROXYSTEROID DEHYDROGENASE TYPE I IN COMPLEX WITH INHIBITOR | | 分子名称: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-[(1R,2s,3S,5s,7s)-5-hydroxytricyclo[3.3.1.1~3,7~]dec-2-yl]-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Branden, G, Boden, C, Ogg, D. | | 登録日 | 2015-03-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design of pyrozolo-pyrimidines as 11beta-HSD1 inhibitors through optimisation of molecular electrostatic potential.
Medchemcomm, 6, 2016
|
|
5BWR
 
 | |
6Y0O
 
 | | isopenicillin N synthase in complex with ACV and Fe under anaerobic environment using FT-SSX methods | | 分子名称: | Anaerobic Fixed Target Structure of Isopenicillin N synthase in complex with Fe and ACV, FE (III) ION, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | 著者 | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | 登録日 | 2020-02-10 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
1I3P
 
 | |
1I3S
 
 | | THE 2.7 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A MUTATED BACULOVIRUS P35 AFTER CASPASE CLEAVAGE | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, EARLY 35 KDA PROTEIN | | 著者 | dela Cruz, W.P, Lemongello, D, Friesen, P.D, Fisher, A.J. | | 登録日 | 2001-02-15 | | 公開日 | 2001-10-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of baculovirus P35 reveals a novel conformational change in the reactive site loop after caspase cleavage.
J.Biol.Chem., 276, 2001
|
|
6Z0O
 
 | | Structure of Affimer-NP bound to Crimean-Congo Haemorrhagic Fever Virus Nucleocapsid Protein | | 分子名称: | Affimer-NP, Nucleocapsid | | 著者 | Alvarez-Rodriguez, B, Tiede, C, Trinh, C, Tomlinson, D, Edwards, T.A, Barr, J.N. | | 登録日 | 2020-05-10 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5974 Å) | | 主引用文献 | Characterization and applications of a Crimean-Congo hemorrhagic fever virus nucleoprotein-specific Affimer: Inhibitory effects in viral replication and development of colorimetric diagnostic tests.
Plos Negl Trop Dis, 14, 2020
|
|
6I0L
 
 | | Crystal structure of human carbonic anhydrase I in complex with the 1-[4-chloro-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea inhibitor | | 分子名称: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea, ACETATE ION, Carbonic anhydrase 1, ... | | 著者 | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | 登録日 | 2018-10-26 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Carbonic anhydrase inhibitors based on sorafenib scaffold: Design, synthesis, crystallographic investigation and effects on primary breast cancer cells.
Eur.J.Med.Chem., 182, 2019
|
|
1JG1
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | 著者 | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | 登録日 | 2001-06-22 | | 公開日 | 2001-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG4
 
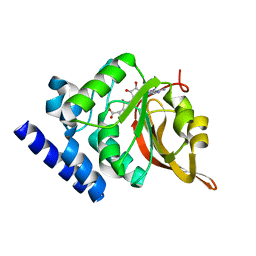 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | 分子名称: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | 著者 | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | 登録日 | 2001-06-22 | | 公開日 | 2001-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JV2
 
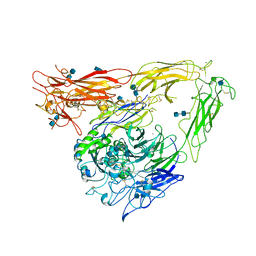 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHAVBETA3 | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xiong, J.P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D, Joachimiak, A, Goodman, S.L, Arnaout, M.A. | | 登録日 | 2001-08-28 | | 公開日 | 2001-10-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of the extracellular segment of integrin alpha Vbeta3.
Science, 294, 2001
|
|
5C7L
 
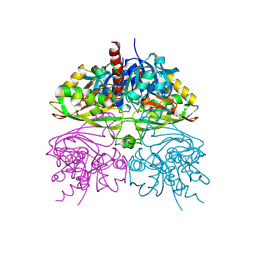 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase apo form | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | | 著者 | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | 登録日 | 2015-06-24 | | 公開日 | 2016-03-09 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.862 Å) | | 主引用文献 | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
6XEB
 
 | |
5AC8
 
 | | S. enterica HisA with mutations D10G, dup13-15, G102A | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | 著者 | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | 登録日 | 2015-08-12 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1KMZ
 
 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | 分子名称: | mitomycin-binding protein | | 著者 | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | 登録日 | 2001-12-17 | | 公開日 | 2002-07-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
4KTO
 
 | | Crystal Structure Of a Putative Isovaleryl-CoA dehydrogenase (PSI-NYSGRC-012251) from Sinorhizobium meliloti 1021 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, isovaleryl-CoA dehydrogenase | | 著者 | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Himmel, D, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-05-20 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.137 Å) | | 主引用文献 | Crystal structure of a Putative Isovaleryl-CoA dehydrogenase from Sinorhizobium meliloti 1021
to be published
|
|
1JG3
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | 分子名称: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | 著者 | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | 登録日 | 2001-06-22 | | 公開日 | 2001-11-16 | | 最終更新日 | 2011-07-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | 分子名称: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | 著者 | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | 登録日 | 2019-04-24 | | 公開日 | 2019-09-11 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
6ZBT
 
 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser342 | | 分子名称: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | 著者 | Joshi, R, Kalabova, D, Obsil, T, Obsilova, V. | | 登録日 | 2020-06-09 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79948521 Å) | | 主引用文献 | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
6ZC9
 
 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser448 | | 分子名称: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | 著者 | Pohl, P, Kalabova, D, Obsil, T, Obsilova, V. | | 登録日 | 2020-06-10 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.89895988 Å) | | 主引用文献 | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
1KLJ
 
 | | Crystal structure of uninhibited factor VIIa | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | 著者 | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | 登録日 | 2001-12-12 | | 公開日 | 2002-10-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
5B1J
 
 | | Crystal structure of the electron-transfer complex of copper nitrite reductase with a cupredoxin | | 分子名称: | Blue copper protein, COPPER (II) ION, Copper-containing nitrite reductase | | 著者 | Nojiri, M, Koteishi, H, Yoneda, R, Hira, D. | | 登録日 | 2015-12-04 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and Function of Copper Nitrite Reductase
Metalloenzymes in denitrification: Applications and Environmental impacts, 2016
|
|
