7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.185 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KOX
 
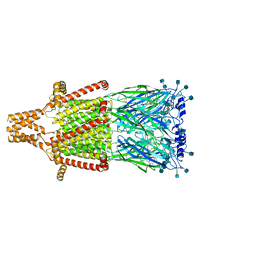 | | Alpha-7 nicotinic acetylcholine receptor bound to epibatidine and PNU-120596 in the activated state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Baxter, L, Cabuco, R, Borek, D, Sine, S. | | 登録日 | 2020-11-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KOO
 
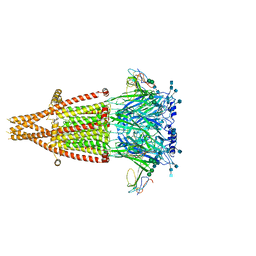 | | Alpha-7 nicotinic acetylcholine receptor bound to alpha-bungarotoxin in a resting state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-bungarotoxin isoform V31, ... | | 著者 | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Cabuco, R, Baxter, L, Borek, D, Sine, S. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KOQ
 
 | | Alpha-7 nicotinic acetylcholine receptor bound to epibatidine in a desensitized state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Cabuco, R, Baxter, L, Borek, D, Sine, S. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KS0
 
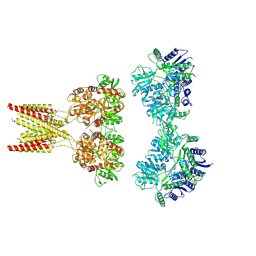 | | GluK2/K5 with 6-Cyano-7-nitroquinoxaline-2,3-dione (CNQX) | | 分子名称: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | 著者 | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | 登録日 | 2020-11-20 | | 公開日 | 2021-03-24 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
7KS3
 
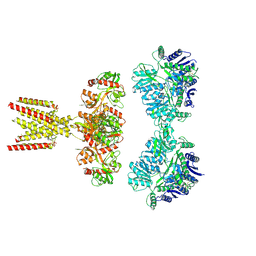 | | GluK2/K5 with L-Glu | | 分子名称: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | 著者 | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | 登録日 | 2020-11-20 | | 公開日 | 2021-03-24 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
3TTI
 
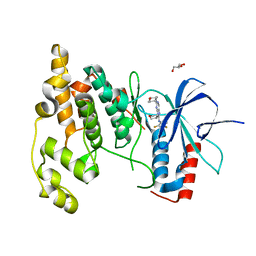 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | 分子名称: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | 著者 | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | 登録日 | 2011-09-14 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3QP5
 
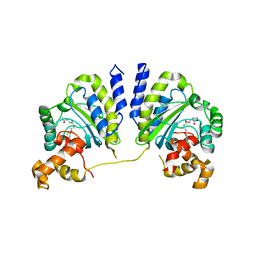 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | 分子名称: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.249 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QUE
 
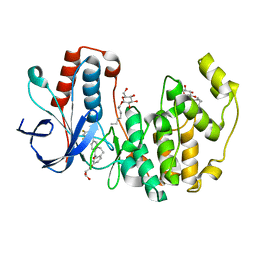 | | Human p38 MAP Kinase in Complex with Skepinone-L | | 分子名称: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | 著者 | Gruetter, C, Mayer-Wrangowski, S, Richters, A, Rauh, D. | | 登録日 | 2011-02-23 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor.
Nat.Chem.Biol., 8, 2012
|
|
3QWY
 
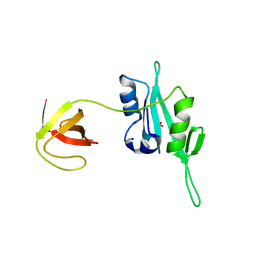 | | CED-2 | | 分子名称: | Cell death abnormality protein 2, GLYCEROL, SULFATE ION | | 著者 | Kang, Y, Sun, J, Liu, Y, Sun, D, Hu, Y, Liu, Y.F. | | 登録日 | 2011-02-28 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
3QU9
 
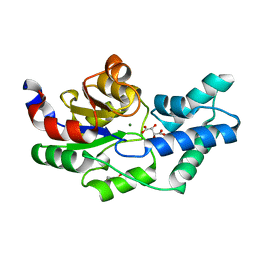 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant complexed with magnesium and tartrate | | 分子名称: | CHLORIDE ION, GLYCEROL, INORGANIC PYROPHOSPHATASE, ... | | 著者 | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | 登録日 | 2011-02-23 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3Q6A
 
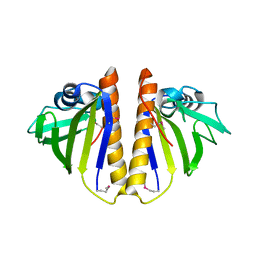 | | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116 | | 分子名称: | uncharacterized protein | | 著者 | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-12-31 | | 公開日 | 2011-04-06 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116
To be Published
|
|
7M5I
 
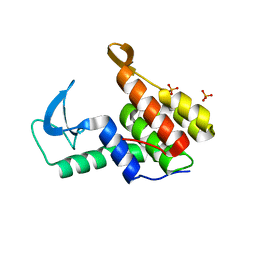 | | Endolysin from Escherichia coli O157:H7 phage FAHEc1 | | 分子名称: | Endolysin, PHOSPHATE ION | | 著者 | Love, M.J, Coombes, D, Billington, C, Dobson, R.C.J. | | 登録日 | 2021-03-24 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | The Molecular Basis for Escherichia coli O157:H7 Phage FAHEc1 Endolysin Function and Protein Engineering to Increase Thermal Stability.
Viruses, 13, 2021
|
|
3QBN
 
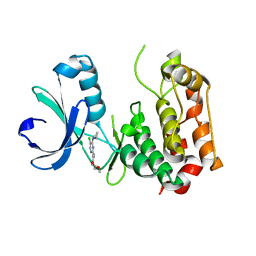 | |
3QJ3
 
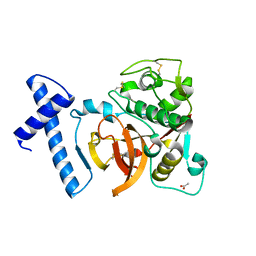 | | Structure of digestive procathepsin L2 proteinase from Tenebrio molitor larval midgut | | 分子名称: | ACETATE ION, Cathepsin L-like protein | | 著者 | Beton, D, Guzzo, C.R, Terra, W.R, Farah, C.S. | | 登録日 | 2011-01-28 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The 3D structure and function of digestive cathepsin L-like proteinases of Tenebrio molitor larval midgut.
Insect Biochem.Mol.Biol., 42, 2012
|
|
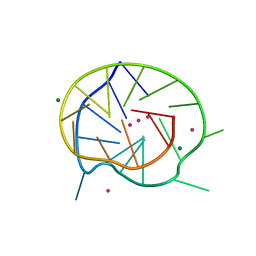
3QXR
 
 | |
3QP6
 
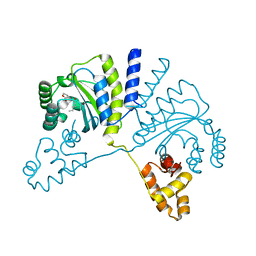 | | Crystal structure of CviR (Chromobacterium violaceum 12472) bound to C6-HSL | | 分子名称: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3Q5K
 
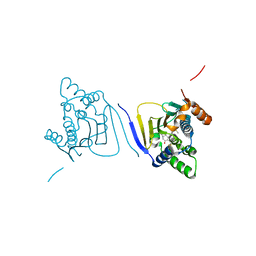 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor | | 分子名称: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-{[2-(methylsulfanyl)ethyl]amino}benzamide, Heat shock protein 83-1 | | 著者 | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-28 | | 公開日 | 2011-02-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor
To be Published
|
|
3Q65
 
 | | Human Aldose Reductase in Complex with NADP+ in Space Group P212121 | | 分子名称: | Aldose reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Sawaya, M.R, Cascio, D, Balendiran, G.K. | | 登録日 | 2010-12-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The role of Cys-298 in aldose reductase function.
J.Biol.Chem., 286, 2011
|
|
7KSQ
 
 | | The Structure of the moss PSI-LHCI reveals the evolution of the LHCI antenna | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Riddle, R, Gorski, C, Toporik, H, Dobson, Z, Da, Z, Williams, D, Mazor, Y. | | 登録日 | 2020-11-23 | | 公開日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4.
Nat.Plants, 8, 2022
|
|
7KU5
 
 | | The Structure of the moss PSI-LHCI reveals the evolution of the LHCI antenna | | 分子名称: | BETA-CAROTENE, CHLOROPHYLL A, PsaO | | 著者 | Riddle, R, Gorski, C, Toporik, H, Dobson, Z, Da, Z, Williams, D, Mazor, Y. | | 登録日 | 2020-11-24 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4.
Nat.Plants, 8, 2022
|
|
