5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
126D
 
 | |
5X9Q
 
 | | Crystal structure of HldC from Burkholderia pseudomallei | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative cytidylyltransferase | | 著者 | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | 登録日 | 2017-03-08 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of D-glycero-Beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei.
Proteins, 86, 2018
|
|
5K89
 
 | | Crystal Structure of Human Calcium-Bound S100A1 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Protein S100-A1 | | 著者 | Melville, Z, Aligholizadeh, E, McKnight, L.E, Weber, D, Pozharski, E, Weber, D.J. | | 登録日 | 2016-05-27 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.249 Å) | | 主引用文献 | X-ray crystal structure of human calcium-bound S100A1.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
131D
 
 | | THE LOW-TEMPERATURE CRYSTAL STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA REVEALS BINDING OF A SPERMINE MOLECULE IN THE MINOR GROOVE | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SODIUM ION, SPERMINE | | 著者 | Bancroft, D, Williams, L.D, Rich, A, Egli, M. | | 登録日 | 1993-06-18 | | 公開日 | 1993-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove.
Biochemistry, 33, 1994
|
|
5K5S
 
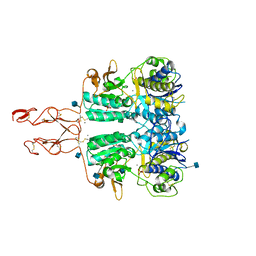 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | 著者 | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | 登録日 | 2016-05-23 | | 公開日 | 2016-08-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
1AJJ
 
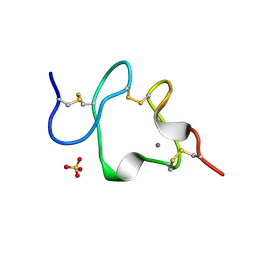 | | LDL RECEPTOR LIGAND-BINDING MODULE 5, CALCIUM-COORDINATING | | 分子名称: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, SULFATE ION | | 著者 | Fass, D, Blacklow, S.C, Kim, P.S, Berger, J.M. | | 登録日 | 1997-05-04 | | 公開日 | 1997-07-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module.
Nature, 388, 1997
|
|
7ACD
 
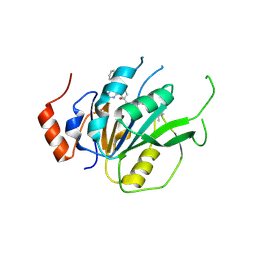 | | Crystal structure of the human METTL3-METTL14 complex with compound T30 (UZH1a) | | 分子名称: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-oxidanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Bedi, R.K, Huang, D, Caflisch, A. | | 登録日 | 2020-09-10 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes.
Chemmedchem, 16, 2021
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | 分子名称: | ZIKV NS1 | | 著者 | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | 登録日 | 2017-03-03 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.817 Å) | | 主引用文献 | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
5XXA
 
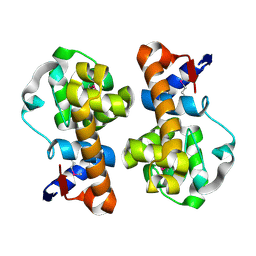 | | beta-1,4-mannanase-SeMet-RmMan134A | | 分子名称: | endo-1,4-beta-mannanase | | 著者 | Jiang, Z.Q, You, X, Yang, D, Huang, P. | | 登録日 | 2017-07-03 | | 公開日 | 2018-07-25 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structure of endo-1,4-beta-mannanase at 1.76 Angstroms resolution.
To Be Published
|
|
1AK8
 
 | | NMR SOLUTION STRUCTURE OF CERIUM-LOADED CALMODULIN AMINO-TERMINAL DOMAIN (CE2-TR1C), 23 STRUCTURES | | 分子名称: | CALMODULIN, CERIUM (III) ION | | 著者 | Bentrop, D, Bertini, I, Cremonini, M.A, Forsen, S, Luchinat, C, Malmendal, A. | | 登録日 | 1997-05-29 | | 公開日 | 1997-09-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the paramagnetic complex of the N-terminal domain of calmodulin with two Ce3+ ions by 1H NMR.
Biochemistry, 36, 1997
|
|
7ZEG
 
 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7GMP | | 分子名称: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | 著者 | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | 登録日 | 2022-03-31 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
5XFF
 
 | |
7ZEE
 
 | | Human cytosolic 5' nucleotidase IIIB | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methylguanosine phosphate-specific 5'-nucleotidase, ... | | 著者 | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | 登録日 | 2022-03-31 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
5K7D
 
 | |
1A3P
 
 | | ROLE OF THE 6-20 DISULFIDE BRIDGE IN THE STRUCTURE AND ACTIVITY OF EPIDERMAL GROWTH FACTOR, NMR, 20 STRUCTURES | | 分子名称: | EPIDERMAL GROWTH FACTOR | | 著者 | Barnham, K, Torres, A, Alewood, D, Alewood, P, Domagala, T, Nice, E, Norton, R. | | 登録日 | 1998-01-22 | | 公開日 | 1998-07-29 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Role of the 6-20 disulfide bridge in the structure and activity of epidermal growth factor.
Protein Sci., 7, 1998
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | 分子名称: | DNA repair and recombination protein RadA, SULFATE ION | | 著者 | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | 登録日 | 2016-06-08 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5KDM
 
 | |
5KD4
 
 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PVI10) of HIV gp120 MN Isolate (IGPGRAFYVI) | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | 著者 | Jiang, J, Natarajan, K, Margulies, D. | | 登録日 | 2016-06-07 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
5XDS
 
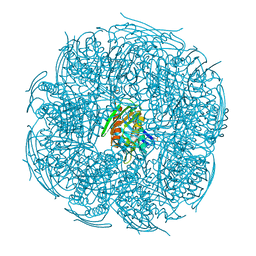 | | Crystal structure of Mycobacterium tuberculosis HisB bound with an inhibitor | | 分子名称: | (2S)-2-azanyl-3-(4H-1,2,4-triazol-3-yl)propanoic acid, CHLORIDE ION, Imidazoleglycerol-phosphate dehydratase, ... | | 著者 | Kumar, D, Jha, B, Ahangar, M.S, Kumar, B.B. | | 登録日 | 2017-03-29 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Characterization of a triazole scaffold compound as an inhibitor of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase.
Proteins, 2021
|
|
5XF2
 
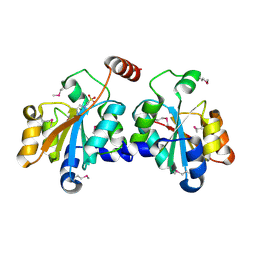 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | 著者 | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | 登録日 | 2017-04-07 | | 公開日 | 2017-07-19 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7ZEH
 
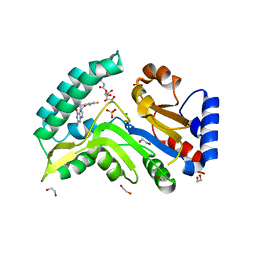 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7Guanine | | 分子名称: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, D-ribulose, ... | | 著者 | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | 登録日 | 2022-03-31 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
5XHI
 
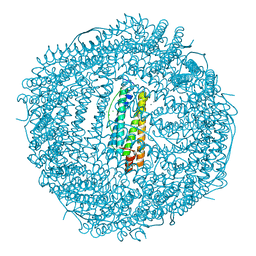 | |
5XI3
 
 | | BRD4 bound with compound Bdi3 | | 分子名称: | (3~{R})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{R})-1-phenylethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | 著者 | Xiong, B, Cao, D, Li, Y. | | 登録日 | 2017-04-25 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.674 Å) | | 主引用文献 | BRD4 bound with compound Bdi3
To Be Published
|
|
5X55
 
 | |
