4YCR
 
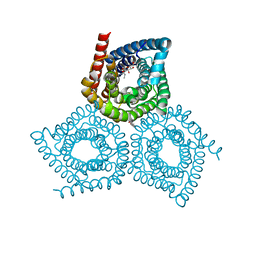 | | Structure determination of an integral membrane protein at room temperature from crystals in situ | | 分子名称: | Tellurite resistance protein TehA homolog, octyl beta-D-glucopyranoside | | 著者 | Axford, D, Hu, N.J, Foadi, J, Choudhury, H.G, Iwata, S, Beis, K, Evans, G, Alguel, Y. | | 登録日 | 2015-02-20 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure determination of an integral membrane protein at room temperature from crystals in situ.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8IKP
 
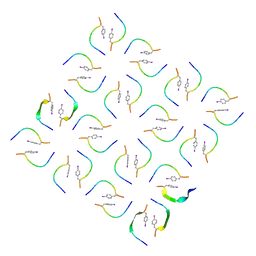 | |
6PVS
 
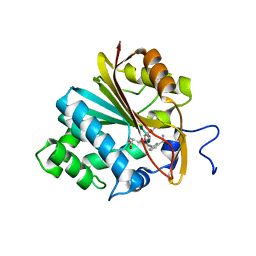 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL320 | | 分子名称: | 9-(5-{[(3R)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)prop-2-yn-1-yl]amino}-5-deoxy-alpha-D-lyxofuranosyl)-9H-purin-6-amine, NNMT protein | | 著者 | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | 登録日 | 2019-07-21 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.575 Å) | | 主引用文献 | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
8IKB
 
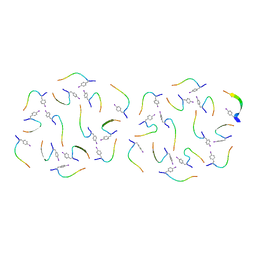 | |
3B1U
 
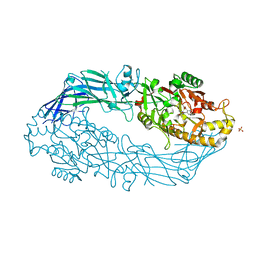 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-F-amidine | | 分子名称: | 2-{[(2S)-1-amino-5-{[(1Z)-2-fluoroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | 著者 | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | 登録日 | 2011-07-13 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
4YIN
 
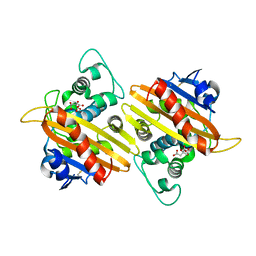 | | Crystal structure of the extended-spectrum beta-lactamase OXA-145 | | 分子名称: | Beta-lactamase, CITRATE ANION | | 著者 | Meziane-Cherif, D, Bonnet, R, Haouz, A, Courvalin, P. | | 登録日 | 2015-03-02 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the loss of penicillinase and the gain of ceftazidimase activities by OXA-145 beta-lactamase in Pseudomonas aeruginosa.
J. Antimicrob. Chemother., 71, 2016
|
|
3FPG
 
 | | Crystal Structure of E81Q mutant of MtNAS | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, Putative uncharacterized protein | | 著者 | Dreyfus, C, Pignol, D, Arnoux, P. | | 登録日 | 2009-01-05 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallographic snapshots of iterative substrate translocations during nicotianamine synthesis in Archaea
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6PYT
 
 | | CryoEM Structure of Pyocin R2 - precontracted - trunk | | 分子名称: | Pyocin sheath PA0622, Pyocin tube PA0623 | | 著者 | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
4YMA
 
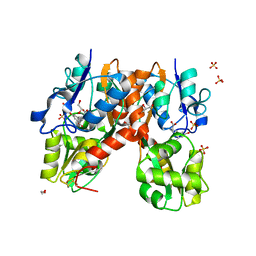 | | Structure of the ligand-binding domain of GluA2 in complex with the antagonist CNG10109 | | 分子名称: | (3R)-3-(3-carboxy-5-hydroxyphenyl)-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | 登録日 | 2015-03-06 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.895 Å) | | 主引用文献 | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
3BIW
 
 | | Crystal structure of the Neuroligin-1/Neurexin-1beta synaptic adhesion complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | 登録日 | 2007-12-01 | | 公開日 | 2007-12-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
6Q84
 
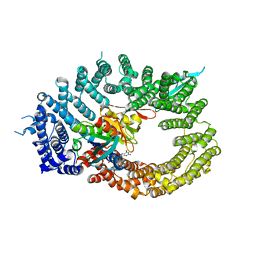 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | 分子名称: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | 登録日 | 2018-12-14 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6Q7F
 
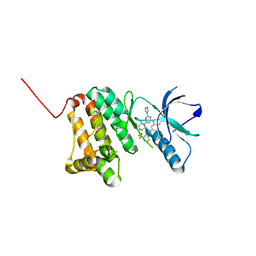 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATDL18 | | 分子名称: | 3-[(4-imidazol-1-yl-6-piperazin-1-yl-1,3,5-triazin-2-yl)amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | 著者 | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | 登録日 | 2018-12-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.204 Å) | | 主引用文献 | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
4YJJ
 
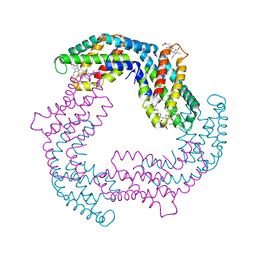 | |
2V8G
 
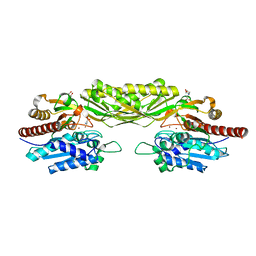 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with the product beta-alanine | | 分子名称: | BETA-ALANINE, BETA-ALANINE SYNTHASE, BICINE, ... | | 著者 | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | 登録日 | 2007-08-07 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
6QA1
 
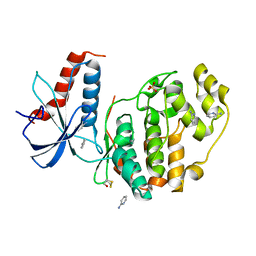 | | ERK2 mini-fragment binding | | 分子名称: | Mitogen-activated protein kinase 1, SULFATE ION, pyridin-2-amine | | 著者 | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | 登録日 | 2018-12-18 | | 公開日 | 2019-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6QAE
 
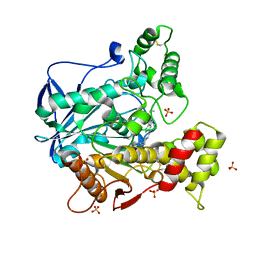 | | Human Butyrylcholinesterase in complex with (S)-N2-butyl-N1-(2-cycloheptylethyl)-3-(1H-indol-3-yl)-N1,N2-dimethylpropane-1,2-diamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | 著者 | Brazzolotto, X, Nachon, F, Harst, M, Knez, D, Gobec, S. | | 登録日 | 2018-12-19 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.487 Å) | | 主引用文献 | Tryptophan-derived butyrylcholinesterase inhibitors as promising leads against Alzheimer's disease.
Chem.Commun.(Camb.), 55, 2019
|
|
4HB5
 
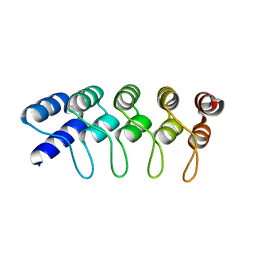 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR267. | | 分子名称: | Engineered Protein | | 著者 | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-09-27 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.294 Å) | | 主引用文献 | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
4YM7
 
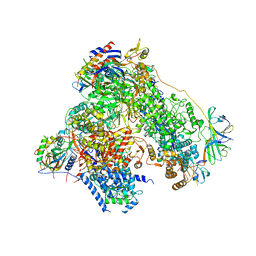 | | RNA polymerase I structure with an alternative dimer hinge | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Kostrewa, D, Kuhn, C.-D, Engel, C, Cramer, P. | | 登録日 | 2015-03-06 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | An alternative RNA polymerase I structure reveals a dimer hinge.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6QBN
 
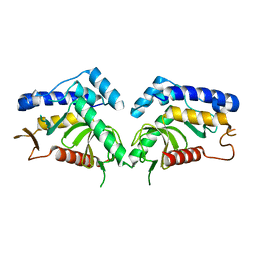 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200D | | 分子名称: | Cell wall assembly regulator SMI1 | | 著者 | Carivenc, C, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | 登録日 | 2018-12-21 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200A
To Be Published
|
|
6PVA
 
 | | The structure of NTMT1 in complex with compound 11 | | 分子名称: | AMINO GROUP-()-LYSINE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | 著者 | Noinaj, N, Chen, D, Huang, R. | | 登録日 | 2019-07-20 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | The structure of NTMT1 in complex with compound 11
To Be published
|
|
3FPH
 
 | |
6PVE
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL319 | | 分子名称: | 9-(5-{[(3S)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)propyl]amino}-5-deoxy-alpha-D-ribofuranosyl)-9H-purin-6-amine, NNMT protein | | 著者 | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | 登録日 | 2019-07-20 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
4YMB
 
 | | Structure of the ligand-binding domain of GluK1 in complex with the antagonist CNG10111 | | 分子名称: | (3R,4S)-3-(3-carboxyphenyl)-4-propyl-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | 登録日 | 2015-03-06 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
3BCV
 
 | |
