2X52
 
 | | CRYSTAL STRUCTURE OF WHEAT GERM AGGLUTININ ISOLECTIN 3 IN COMPLEX WITH A SYNTHETIC DIVALENT CARBOHYDRATE LIGAND | | Descriptor: | AGGLUTININ ISOLECTIN 3, BIS-(2-ACETAMIDO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYLOXYCARBONYL)-4,7,10-TRIOXA-1,13-TRIDECANEDIAMINE, GLYCEROL | | Authors: | Schwefel, D, Maierhofer, C, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
8AFW
 
 | | Tube assembly of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFY
 
 | | Subtomogram average of membrane-bound Atg18 oligomers | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8ALM
 
 | |
8AHY
 
 | |
8B20
 
 | | NDM-1 metallo-beta-lactamase in complex with triazole-based inhibitor CP57 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(~{E})-[3-sulfanyl-5-[4-(trifluoromethyl)phenyl]-1,2,4-triazol-4-yl]iminomethyl]benzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Lazzarato, L, Bersani, M, Gianquinto, E, Docquier, J.D, Spyrakis, F, Tondi, D, Cendron, L. | | Deposit date: | 2022-09-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Optimization of 1,2,4-Triazole-3-Thione Derivatives: Improving Inhibition of NDM-/VIM-Type Metallo-beta-Lactamases and Synergistic Activity on Resistant Bacteria.
Pharmaceuticals, 16, 2023
|
|
1PKW
 
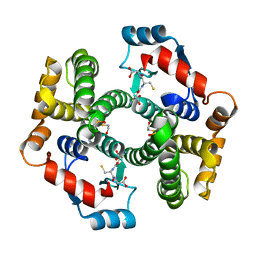 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with glutathione | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8ALH
 
 | |
8B1Z
 
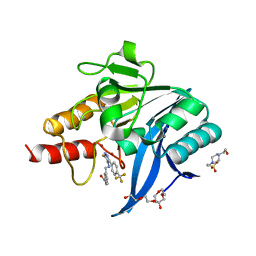 | | NDM-1 metallo-beta-lactamase in complex with triazole-based inhibitor CP56 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[3-sulfanyl-5-[4-(trifluoromethyl)phenyl]-1,2,4-triazol-4-yl]iminomethyl]benzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Lazzarato, L, Bersani, M, Gianquinto, E, Docquier, J.D, Spyrakis, F, Tondi, D, Cendron, L. | | Deposit date: | 2022-09-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of 1,2,4-Triazole-3-Thione Derivatives: Improving Inhibition of NDM-/VIM-Type Metallo-beta-Lactamases and Synergistic Activity on Resistant Bacteria.
Pharmaceuticals, 16, 2023
|
|
8B1W
 
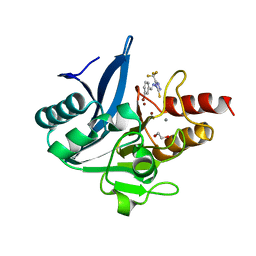 | | NDM-1 metallo-beta-lactamase in complex with triazole-based inhibitor CP35 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[(~{E})-(3-bromophenyl)methylideneamino]-5-(trifluoromethyl)-1,2,4-triazole-3-thiol, ... | | Authors: | Vascon, F, Lazzarato, L, Bersani, M, Gianquinto, E, Docquier, J.D, Spyrakis, F, Tondi, D, Cendron, L. | | Deposit date: | 2022-09-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Optimization of 1,2,4-Triazole-3-Thione Derivatives: Improving Inhibition of NDM-/VIM-Type Metallo-beta-Lactamases and Synergistic Activity on Resistant Bacteria.
Pharmaceuticals, 16, 2023
|
|
2WHS
 
 | | Fluorescent Protein mKeima at pH 3.8 | | Descriptor: | LARGE STOKES SHIFT FLUORESCENT PROTEIN, SULFATE ION | | Authors: | Violot, S, Carpentier, P, Blanchoin, L, Bourgeois, D. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reverse Ph-Dependence of Chromophore Protonation Explains the Large Stokes Shift of the Red Fluorescent Protein Mkeima.
J.Am.Chem.Soc., 131, 2009
|
|
7P06
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P03
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | Descriptor: | Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
8AFX
 
 | | Single particle structure of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures of Atg18 oligomers reveal a tilted structural scaffold for Atg2 at the isolation membrane
To Be Published
|
|
1PT0
 
 | | Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase with an Alanine insertion at position 26 | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-22 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PUZ
 
 | | Solution NMR Structure of Protein NMA1147 from Neisseria meningitidis. Northeast Structural Genomics Consortium Target MR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Liu, G, Xu, D, Sukumaran, D.K, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-25 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the hypothetical protein NMA1147 from Neisseria meningitidis reveals a distinct 5-helix bundle.
Proteins, 55, 2004
|
|
2WYG
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1R)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2JB3
 
 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with o-aminobenzoate | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
1PWA
 
 | | Crystal structure of Fibroblast Growth Factor 19 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fibroblast growth factor-19, GLYCEROL, ... | | Authors: | Harmer, N.J, Pellegrini, L, Chirgadze, D, Fernandez-Recio, J, Blundell, T.L. | | Deposit date: | 2003-07-01 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of fibroblast growth factor (FGF) 19 reveals novel features of the FGF family and offers a structural basis for its unusual receptor affinity.
Biochemistry, 43, 2004
|
|
7PBK
 
 | | Vibriophage phiVC8 family A DNA polymerase (DpoZ), two conformations: thumb-exo open and thumb-exo closed | | Descriptor: | DNA polymerase I | | Authors: | Czernecki, D, Hu, H, Romoli, F, Delarue, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage phi VC8.
Nucleic Acids Res., 49, 2021
|
|
7P47
 
 | | Structure of the E3 ligase Smc5/Nse2 in complex with Ubc9-SUMO thioester mimetic | | Descriptor: | E3 SUMO-protein ligase MMS21, SUMO-conjugating enzyme UBC9, Structural maintenance of chromosomes protein 5, ... | | Authors: | Lascorz, J, Varejao, N, Reverter, D. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.314 Å) | | Cite: | Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs.
Nat Commun, 12, 2021
|
|
