8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
2M5W
 
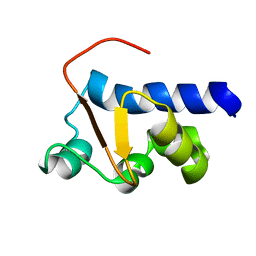 | | NMR Solution Structure of the La motif (N-terminal Domain, NTD) of Dictyostelium discoideum La protein | | Descriptor: | Lupus La protein | | Authors: | Vourtsis, D.J, Chasapis, C.T, Apostolidi, M, Stathopoulos, C, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the La motif (N-terminal Domain, NTD) of Dictyostelium discoideum La protein
To be Published
|
|
8FFE
 
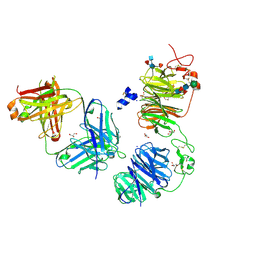 | | Crystal structure of LRP6 E1E2 domains bound to YW210.09 Fab and engineered XWnt8 peptide | | Descriptor: | GLYCEROL, Low-density lipoprotein receptor-related protein 6, SODIUM ION, ... | | Authors: | Jude, K.M, Tsutsumi, N, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FWI
 
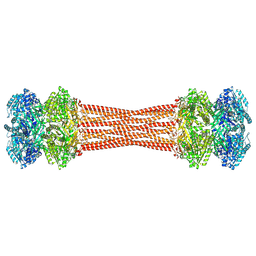 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWJ
 
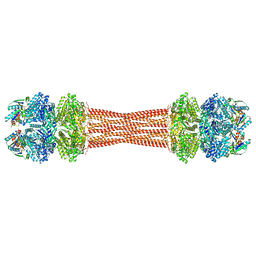 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
1K56
 
 | | OXA 10 class D beta-lactamase at pH 6.5 | | Descriptor: | OXA10 beta-lactamase, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2M68
 
 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R in complex with IGF2 (domain 11 structure only) | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | Deposit date: | 2013-03-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1K57
 
 | | OXA 10 class D beta-lactamase at pH 6.0 | | Descriptor: | BETA LACTAMASE OXA-10, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2J6P
 
 | | STRUCTURE OF AS-SB REDUCTASE FROM LEISHMANIA MAJOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SB(V)-AS(V) REDUCTASE, ... | | Authors: | Bisacchi, D, Zhou, Y, Rosen, B.P, Mukhopadhyay, R, Bordo, D. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-02 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the As/Sb reductase LmACR2 from Leishmania major.
J. Mol. Biol., 386, 2009
|
|
8FLU
 
 | | Human PTH1R in complex with LA-PTH and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
6SFR
 
 | | SOS1 in Complex with Inhibitor BI-68BS | | Descriptor: | 6,7-dimethoxy-~{N}-[(1~{R})-1-phenylethyl]quinazolin-4-amine, IMIDAZOLE, Son of sevenless homolog 1 | | Authors: | Kessler, D, Fischer, G, Ramharter, J. | | Deposit date: | 2019-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition.
Cancer Discov, 11, 2021
|
|
8FLR
 
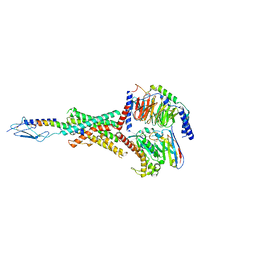 | | Human PTH1R in complex with PTHrP and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
2JS5
 
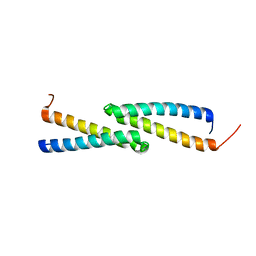 | | NMR Structure of protein Q60C73_METCA. Northeast Structural Genomics Consortium target McR1 | | Descriptor: | Uncharacterized protein | | Authors: | Singarapu, K.K, Wu, Y, Eletsky, A, Sukumaran, D, Parish, D, Chen, C.X, Nwosu, C, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of protein Q60C73_METCA.
To be Published
|
|
8FLT
 
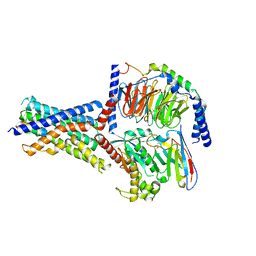 | | Human PTH1R in complex with M-PTH(1-14) and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
8FLQ
 
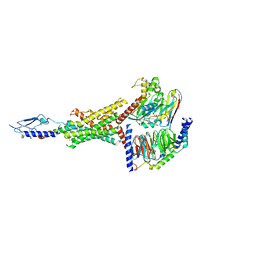 | | Human PTH1R in complex with PTH(1-34) and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
5M3S
 
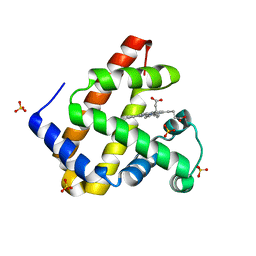 | | Low-dose fixed target serial synchrotron crystallography structure of Metmyoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Axford, D, Owen, R.L, Sherrell, D, Muller-Werkmeister, H. | | Deposit date: | 2016-10-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low-dose fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8FLS
 
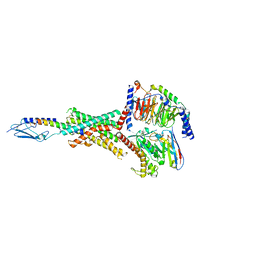 | | Human PTH1R in complex with Abaloparatide and Gs | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
6AU2
 
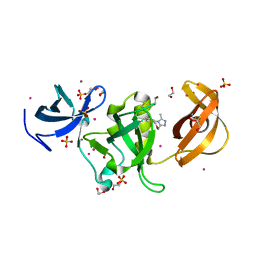 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-10-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
6SC9
 
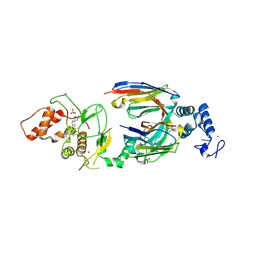 | | dAb3/HOIP-RBR-HOIPIN-8 | | Descriptor: | 2-[3-[2,6-bis(fluoranyl)-4-(1~{H}-pyrazol-4-yl)phenyl]-3-oxidanylidene-prop-1-enyl]-4-(1-methylpyrazol-4-yl)benzoic acid, CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
6SAK
 
 | |
7GH9
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-362d364a-10 (Mpro-x2971) | | Descriptor: | 2-[(2S)-2-{2-[(methanesulfonyl)amino]ethyl}piperidin-1-yl]-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHG
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-4 (Mpro-x3324) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-1-(3-chlorophenyl)-2-hydroxyethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ADA-UNI-f8e79267-2 (Mpro-x10889) | | Descriptor: | (2R)-4-[(methanesulfonyl)amino]-2-phenyl-N-(pyridin-3-yl)butanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TOB-UNK-c2aba166-1 (Mpro-x3325) | | Descriptor: | 1-[4-(prop-2-yn-1-yl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
