6Z6W
 
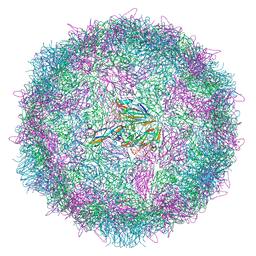 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) from a mammalian expression system. | | Descriptor: | Capsid proteins, VP0, VP1, ... | | Authors: | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mammalian expression of virus-like particles as a proof of principle for next generation polio vaccines.
NPJ Vaccines, 6, 2021
|
|
7TII
 
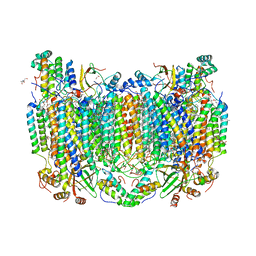 | | Annealed structure of oxidized bovine cytochrome c oxidase with reduced metal centers induced by synchrotron X-ray exposure | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R, Russi, S, Cohen, A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
4YEU
 
 | |
7QYY
 
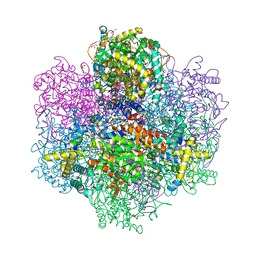 | | Vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with chloride | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-29 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
6NR8
 
 | | hTRiC-hPFD Class6 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
8A3J
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil heterotetramer with 3 heptad repeats, apCC-Tet*3-A2B2 | | Descriptor: | apCC-Tet*3-A, apCC-Tet*3-B | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
7TIH
 
 | | Structure of oxidized bovine cytochrome c oxidase with reduced metal centers induced by synchrotron X-ray exposure | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R, Russi, S, Cohen, A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
5FRO
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', 5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP *AP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
4YJS
 
 | | THE KINASE DOMAIN OF HUMAN SPLEEN TYROSINE (SYK) IN COMPLEX WITH GTC000226 | | Descriptor: | 3-[{2-[(1,1-dioxido-2,3-dihydro-1,2-benzothiazol-6-yl)amino]pyrimidin-4-yl}(1H-indazol-4-yl)amino]propan-1-ol, Tyrosine-protein kinase SYK | | Authors: | Somers, D.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of GSK143, a highly potent, selective and orally efficacious spleen tyrosine kinase inhibitor.
Bioorg. Med. Chem. Lett., 21, 2011
|
|
5KRS
 
 | | HIV-1 Integrase Catalytic Core Domain in Complex with an Allosteric Inhibitor, 3-(1H-pyrrol-1-yl)-2-thiophenecarboxylic acid | | Descriptor: | 3-pyrrol-1-ylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, Integrase | | Authors: | Patel, D, Bauman, J.D, Arnold, E. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-28 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
J.Biol.Chem., 291, 2016
|
|
5A0C
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | (6S)-6-(4-cyano-2-methylsulfonyl-phenyl)-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-5-carbonitrile, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
5LCK
 
 | | A Clickable Covalent ERK 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, ~{N}-[2-[[2-[(5-methoxypyridin-3-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]propanamide | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
5KMH
 
 | | Structure of CavAb in complex with Br-verapamil | | Descriptor: | (2~{R})-2-(2-bromophenyl)-5-[2-(3,4-dimethoxyphenyl)ethyl-methyl-amino]-2-propan-2-yl-pentanenitrile, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
7QAR
 
 | | Serial crystallography structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-methyl uric acid at room temperature | | Descriptor: | (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, Uricase | | Authors: | Bui, S, Catapano, L, Zielinski, K, Yefanov, O, Murshudov, G.N, Oberthuer, D, Steiner, R.A. | | Deposit date: | 2021-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
5KNT
 
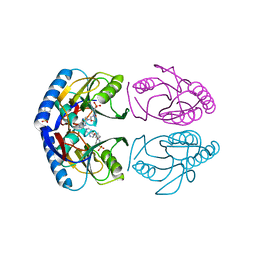 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with 2-((2,3-Dihydroxypropyl)(2-(hypoxanthin-9-yl)ethyl)amino)ethylphosphonic acid | | Descriptor: | 2-[[(2~{S})-2,3-bis(oxidanyl)propyl]-[2-(6-oxidanylidene-1~{H}-purin-9-yl)ethyl]amino]ethylphosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNV
 
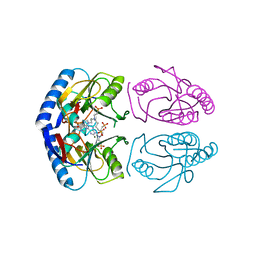 | | E coli hypoxanthine guanine phosphoribosyltransferase in complexed with 9-[(N-Phosphonoethyl-N-phosphonobutyl)-2-aminoethyl]-hypoxanthine | | Descriptor: | 2-[2-(6-oxidanylidene-1~{H}-purin-9-yl)ethyl-(4-phosphonobutyl)amino]ethylphosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
6X1K
 
 | | Solution NMR structure of de novo designed TMB2.3 | | Descriptor: | De novo designed transmembrane beta-barrel TMB2.3 | | Authors: | Liang, B, Vorobieva, A.A, Chow, C.M, Baker, D, Tamm, L.K. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of transmembrane beta barrels.
Science, 371, 2021
|
|
7LQV
 
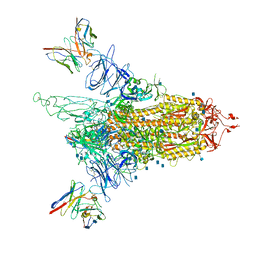 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
5N6G
 
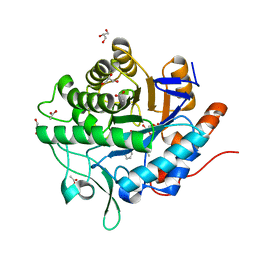 | | NerA from Agrobacterium radiobacter in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
8HWQ
 
 | |
8VEX
 
 | |
8VEY
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and TNG908 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
7TPR
 
 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
7ODC
 
 | | CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kern, A.D, Oliveira, M.A, Coffino, P, Hackert, M.L. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
Structure Fold.Des., 7, 1999
|
|
6SRX
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
