6U5J
 
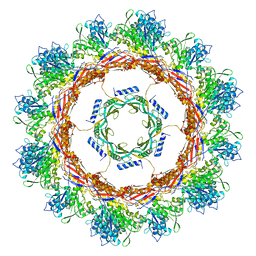 | | CryoEM Structure of Pyocin R2 - postcontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
8BWV
 
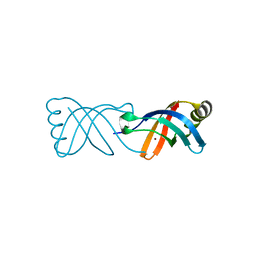 | |
5IEY
 
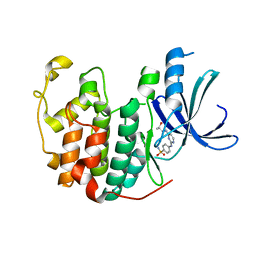 | | Crystal structure of a CDK inhibitor bound to CDK2 | | Descriptor: | 4-[(4-{[(2R,3R)-3-hydroxybutan-2-yl]amino}pyrimidin-2-yl)amino]benzene-1-sulfonamide, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
7SC0
 
 | |
7SCX
 
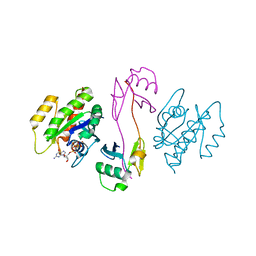 | | KRAS full-length G12V in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
8BTN
 
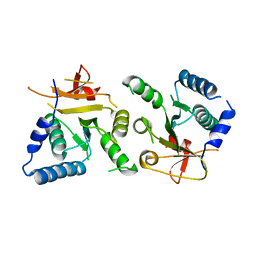 | | Crystal structure of BcThsB | | Descriptor: | TIR domain-containing protein | | Authors: | Tamulaitiene, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
6U7P
 
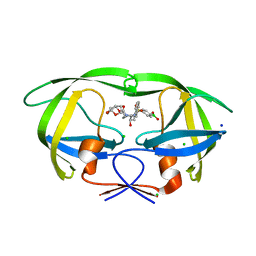 | | HIV-1 wild type protease with GRL-03119A, with phenyl-boronic-acid as P2'-ligand and with a hexahydro-4H-furo-pyran as the P2-ligand | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Kneller, D.W, Weber, I.T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Potent HIV-1 Protease Inhibitors Containing Carboxylic and Boronic Acids: Effect on Enzyme Inhibition and Antiviral Activity and Protein-Ligand X-ray Structural Studies.
Chemmedchem, 14, 2019
|
|
5IG9
 
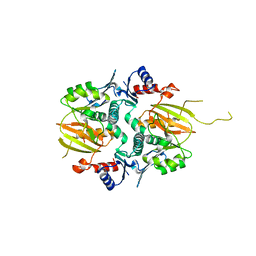 | |
4XBT
 
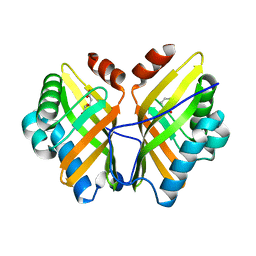 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH complexed with (S,S)-cyclohexanediol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, CITRATE ANION, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7JMN
 
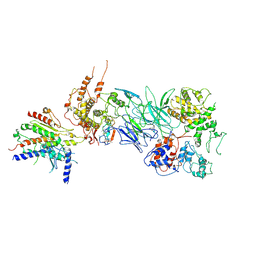 | | Tail module of Mediator complex | | Descriptor: | MED15, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Zhang, H.Q, Chen, D.C. | | Deposit date: | 2020-08-02 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
4XDV
 
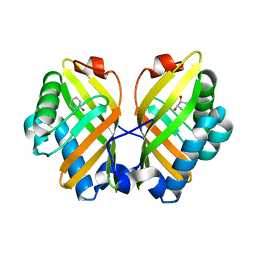 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclohexanediol | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8G5N
 
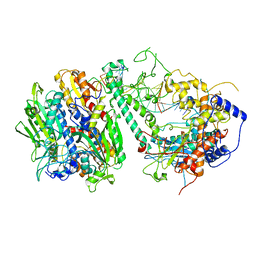 | | Cryo-EM structure of the Guide loop Engagement Complex (VI) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
5INF
 
 | | Structural basis for acyl-CoA carboxylase-mediated assembly of unusual polyketide synthase extender units incorporated into the stambomycin antibiotics | | Descriptor: | Carboxyl transferase, HEXANOYL-COENZYME A | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Withall, D.M, Song, L, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
7SG5
 
 | | Structure of PfCSP peptide 21 with antibody CIS43_Var2 | | Descriptor: | CIS43_Var2 Fab Heavy chain, CIS43_Var2 Fab Light chain, GLYCEROL, ... | | Authors: | Tripathi, P, Kwong, P.D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Highly protective antimalarial antibodies via precision library generation and yeast display screening.
J.Exp.Med., 219, 2022
|
|
8G5K
 
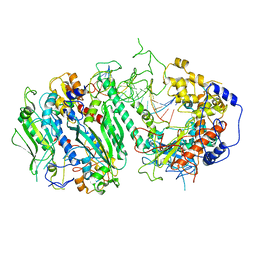 | | Cryo-EM structure of the Wedge Alignment Complex (VIII) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
7SG6
 
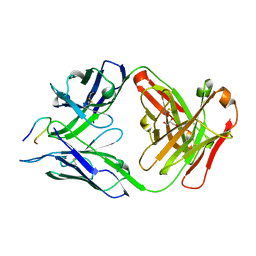 | | Structure of PfCSP peptide 21 with antibody CIS43_Var10 | | Descriptor: | CIS43_Var10 Fab Heavy chain, CIS43_Var10 Fab Light chain, GLYCEROL, ... | | Authors: | Tripathi, P, Kwong, P.D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly protective antimalarial antibodies via precision library generation and yeast display screening.
J.Exp.Med., 219, 2022
|
|
8G5I
 
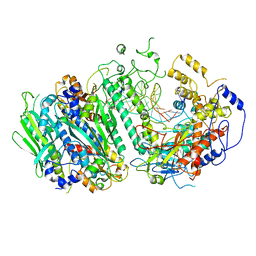 | | Cryo-EM structure of the Mismatch Sensing Complex (I) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8G5M
 
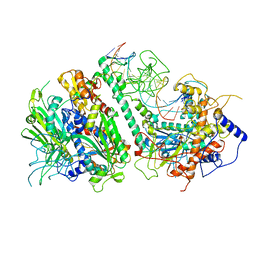 | | Cryo-EM structure of the Mismatch Locking Complex (III) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8G5J
 
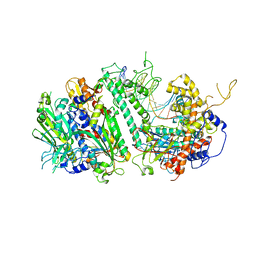 | | Cryo-EM structure of the Mismatch Uncoupling Complex (II) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8G5L
 
 | | Cryo-EM structure of the Primer Separation Complex (IX) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
8G5P
 
 | | Cryo-EM structure of the Guide loop Engagement Complex (V) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
3H40
 
 | | Binary complex of human DNA polymerase iota with template U/T | | Descriptor: | 5'-D(*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(*TP*(BRU)P*GP*GP*GP*TP*CP*CP*T)-3', DNA polymerase iota, ... | | Authors: | Jain, R, Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Replication across template T/U by human DNA polymerase-iota.
Structure, 17, 2009
|
|
8G5O
 
 | | Cryo-EM structure of the Guide loop Engagement Complex (IV) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
7RU6
 
 | |
5IQ3
 
 | |
