2QVZ
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303A mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
2A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
2QX3
 
 | |
2B4V
 
 | | Structural Basis for UTP Specificity of RNA Editing TUTases From Trypanosoma Brucei | | Descriptor: | POTASSIUM ION, RNA editing complex protein MP57 | | Authors: | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
2B61
 
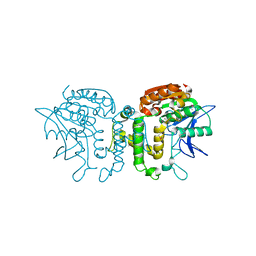 | | Crystal Structure of Homoserine Transacetylase | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Mirza, I.A, Nazi, I, Korczynska, M, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Homoserine Transacetylase from Haemophilus influenzae Reveals a New Family of alpha/beta-Hydrolases
Biochemistry, 44, 2005
|
|
2GTH
 
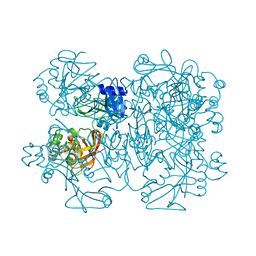 | | crystal structure of the wildtype MHV coronavirus non-structural protein nsp15 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Xu, X, Zhai, Y, Sun, F, Lou, Z, Su, D, Rao, Z. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15
J.Virol., 80, 2006
|
|
2QJ0
 
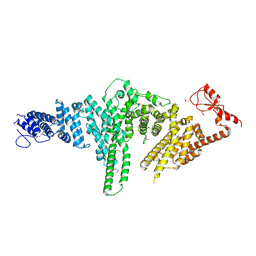 | |
2QJ1
 
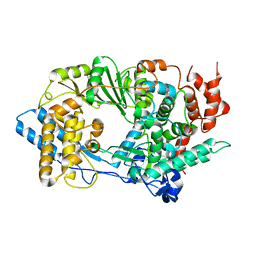 | | Crystal structure of infectious bursal disease virus VP1 polymerase incubated with an oligopeptide mimicking the VP3 C-terminus | | Descriptor: | Infectious bursal disease virus VP1 polymerase | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-07-06 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Unprecedented activation mechanism of a non-canonical RNA-dependent RNA polymerase
To be Published
|
|
2BBI
 
 | |
2QVL
 
 | |
2QVY
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303G mutation, bound to 3,4-Dichlorobenzoate | | Descriptor: | 3,4-dichlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
2HAH
 
 | | The structure of FIV 12S protease in complex with TL-3 | | Descriptor: | Protease, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Lin, Y.C, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an FIV/HIV chimeric protease complexed with the broad-based inhibitor, TL-3.
Retrovirology, 4, 2007
|
|
2R88
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant delta S3 kink | | Descriptor: | Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane passage of hydrophobic compounds through a protein channel wall
Nature, 458, 2009
|
|
2R70
 
 | | Crystal structure of infectious bursal disease virus VP1 polymerase, cocrystallized with an oligopeptide mimicking the VP3 C-terminus. | | Descriptor: | INFECTIOUS BURSAL VIRUS VP1 POLYMERASE | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-09-07 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R89
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant delta N3 | | Descriptor: | Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | From the Cover: Ligand-gated diffusion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1S9K
 
 | |
7LCI
 
 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
6Z36
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2118 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-(4-piperidin-4-ylphenyl)-5-(3,4,5-trimethoxyphenyl)pyridine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-05-19 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2118
To Be Published
|
|
1SFH
 
 | | Reduced state of amicyanin mutant P94F | | Descriptor: | Amicyanin, COPPER (I) ION, SODIUM ION | | Authors: | Carrell, C.J, Sun, D, Jiang, S, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Studies of Two Mutants of Amicyanin from Paracoccus denitrificans That Stabilize the Reduced State of the Copper.
Biochemistry, 43, 2004
|
|
7LC3
 
 | | CryoEM Structure of KdpFABC in E1-ATP state | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1SIF
 
 | | Crystal structure of a multiple hydrophobic core mutant of ubiquitin | | Descriptor: | ubiquitin | | Authors: | Benitez-Cardoza, C.G, Stott, K, Hirshberg, M, Went, H.M, Woolfson, D.N, Jackson, S.E. | | Deposit date: | 2004-02-29 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exploring sequence/folding space: folding studies on multiple hydrophobic core mutants of ubiquitin
Biochemistry, 43, 2004
|
|
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
6WHB
 
 | | MEKK1 TOG domain (548-867) | | Descriptor: | ACETATE ION, GLYCEROL, Mitogen-activated protein kinase kinase kinase 1 | | Authors: | Filipcik, P, Mace, P.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.90032053 Å) | | Cite: | A cryptic tubulin-binding domain links MEKK1 to curved tubulin protomers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1SJ8
 
 | | Solution Structure of the R1R2 Domains of Talin | | Descriptor: | Talin 1 | | Authors: | Papagrigoriou, E, Gingras, A.R, Barsukov, I.L, Critchley, D.R, Emsley, J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle
EMBO J., 23, 2004
|
|
4BKN
 
 | | Human Dihydropyrimidinase-related protein 3 (DPYSL3) | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 3 | | Authors: | Mathea, S, Elkins, J.M, Alegre-Abarrategui, J, Shrestha, L, Burgess-Brown, N, Puranik, S, Coutandin, D, Bradley, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Dpysl3
To be Published
|
|
