6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
1D67
 
 | | THE MOLECULAR STRUCTURE OF AN IDARUBICIN-D(TGATCA) COMPLEX AT HIGH RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), IDARUBICIN | | Authors: | Gallois, B, Langlois D'Estaintot, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-03-31 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an idarubicin-d(TGATCA) complex at high resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
1D9E
 
 | | STRUCTURE OF E. COLI KDO8P SYNTHASE | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Radaev, S, Dastidar, P, Patel, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 1999-10-27 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of 3-deoxy-D-manno-octulosonate 8-phosphate synthase.
J.Biol.Chem., 275, 2000
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
2PE9
 
 | |
8C5I
 
 | | Cyanide dihydratase from Bacillus pumilus C1 variant - Q86R,H305K,H308K,H323K | | Descriptor: | Cyanide dihydratase | | Authors: | Mulelu, A.E, Reitz, J, van Rooyen, J, Scheffer, M, Frangakis, A.S, Dlamini, L.S, Woodward, J.D, Benedik, M.J, Sewell, B.T. | | Deposit date: | 2023-01-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Role of Histidine Residues in the Oligomerization of Cyanide Dihydratase from Bacillus pumilus C1
To Be Published
|
|
2P9P
 
 | |
8BZV
 
 | |
2PAU
 
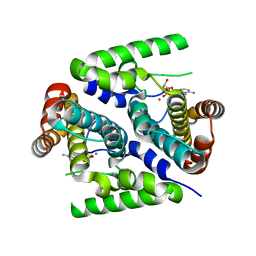 | | Crystal structure of the 5'-deoxynucleotidase YfbR mutant E72A complexed with Co(2+) and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-deoxynucleotidase YfbR, COBALT (II) ION, ... | | Authors: | Zimmerman, M.D, Proudfoot, M, Yakunin, A, Minor, W. | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli.
J.Mol.Biol., 378, 2008
|
|
6YUR
 
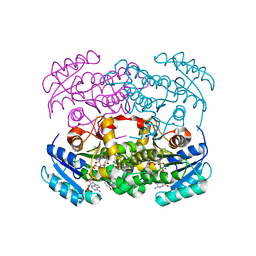 | | Crystal structure of S. aureus FabI inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Weinrich, J.D, Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
1KG6
 
 | | Crystal structure of the K142R mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
8BVZ
 
 | |
2OWQ
 
 | |
8C4I
 
 | |
8C5M
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Tubercidin and Adenosine bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
2OPZ
 
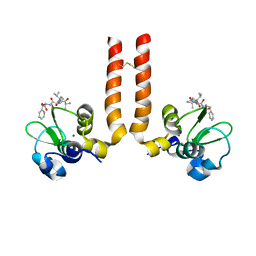 | | AVPF bound to BIR3-XIAP | | Descriptor: | AVPF (Smac homologue, N-terminal tetrapeptide), Baculoviral IAP repeat-containing protein 4, ... | | Authors: | Wist, A.D. | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP.
Bioorg.Med.Chem., 15, 2007
|
|
5JYV
 
 | |
7SZ7
 
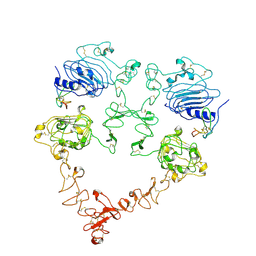 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
1KG4
 
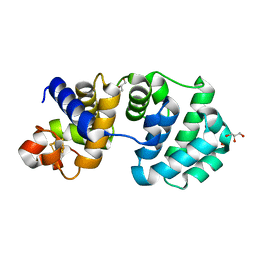 | | Crystal structure of the K142A mutant of E. coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
7SZ1
 
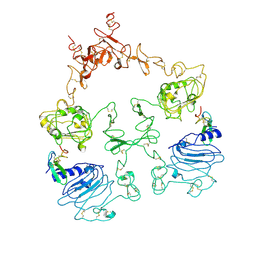 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ0
 
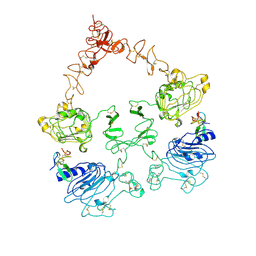 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYD
 
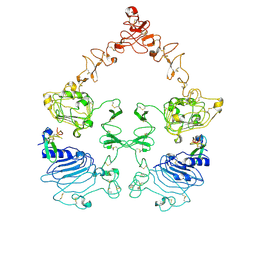 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ5
 
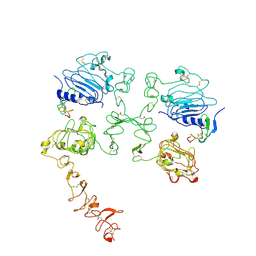 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to TGF-alpha "tips-separated" conformation | | Descriptor: | Epidermal growth factor receptor, Transforming growth factor alpha | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYE
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
