1IMW
 
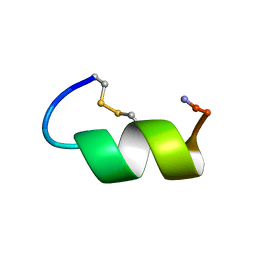 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
8QT0
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-3 | | Descriptor: | 1,2-ETHANEDIOL, 3-dodecylsulfanyl-3-methyl-butanoic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
8QT2
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-6 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
7RU6
 
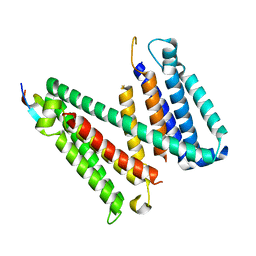 | |
8QOO
 
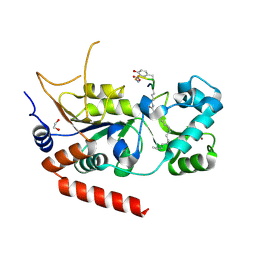 | | Crystal structure of human Sirt2 in complex with the peptide-based pseudo-inhibitor TNFn-4.1 | | Descriptor: | (2S,3R)-3-dodecylsulfanyl-2-methyl-butanoic acid, (2S,3S)-3-dodecylsulfanyl-2-methyl-butanoic acid, (R,R)-2,3-BUTANEDIOL, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of potent TNFa-Myr-based pseudo-inhibitors for Sirtuin 2
To Be Published
|
|
9B53
 
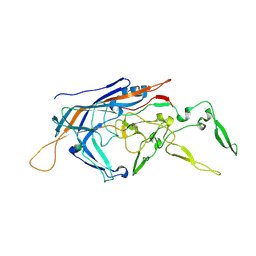 | | RhAAV4282 Full Capsid | | Descriptor: | Capsid protein VP1 | | Authors: | Dagotto, G, Jenni, S, Li, Z, Barouch, D.H. | | Deposit date: | 2024-03-22 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Identification of a novel neutralization epitope in rhesus AAVs.
Mol Ther Methods Clin Dev, 32, 2024
|
|
5HYH
 
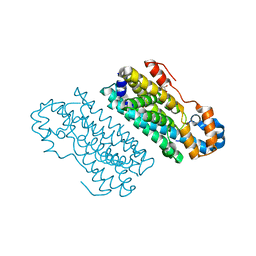 | |
7Z7D
 
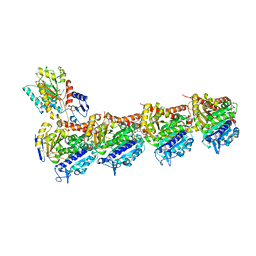 | | Tubulin-Todalam-Vinblastine-complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Roy, B, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7N00
 
 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 648-1025 in complex with AUG-alpha | | Descriptor: | ALK and LTK ligand 2, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Myasnikov, A.G, Rossi, P, Miller, D.J, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
8REJ
 
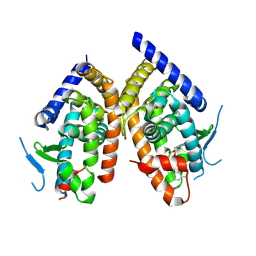 | | Crystal structure of PPAR gamma Ligand Binding Domain in complex with the ligand LBB78 | | Descriptor: | (2~{S})-2-(4-naphthalen-1-ylphenoxy)-3-phenyl-propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Capelli, D, Montanari, R. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | A chemical modification of a peroxisome proliferator-activated receptor pan agonist produced a shift to a new dual alpha/gamma partial agonist endowed with mitochondrial pyruvate carrier inhibition and antidiabetic properties.
Eur.J.Med.Chem., 275, 2024
|
|
6PVB
 
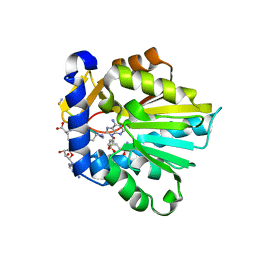 | | The structure of NTMT1 in complex with compound 6 | | Descriptor: | AMINO GROUP-()-(2~{S})-2-azanylpropanal-()-ISOLEUCINE-()-ARGININE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-9-(5-{[(3S)-3-amino-3-carboxypropyl](pentyl)amino}-5-deoxy-beta-L-arabinofuranosyl)-9H-purin-6-amine, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
4X2K
 
 | | Selection of fragments for kinase inhibitor design: decoration is key | | Descriptor: | 4-[(3-aminophenyl)amino]pyrido[2,3-d]pyrimidin-5(6H)-one, SULFATE ION, TGF-beta receptor type-1 | | Authors: | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | Deposit date: | 2014-11-26 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
7RV8
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-10268 | | Descriptor: | DIMETHYL SULFOXIDE, Isoform 2 of B-cell lymphoma 6 protein, N-[5-chloro-2-(morpholin-4-yl)pyridin-4-yl]-2-{5-(3-cyano-4-hydroxy-5-methylphenyl)-3-[3-(1-methyl-1H-pyrazol-4-yl)prop-2-yn-1-yl]-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl}acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the BCL6 BTB domain
To Be Published
|
|
7ND2
 
 | | Cryo-EM structure of the human FERRY complex | | Descriptor: | Glutamine amidotransferase-like class 1 domain-containing protein 1, Protein phosphatase 1 regulatory subunit 21, Quinone oxidoreductase-like protein 1 | | Authors: | Quentin, D, Klink, B.U, Raunser, S. | | Deposit date: | 2021-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
8QT8
 
 | | Crystal structure of human Sirt2 in complex with a TNFa-Myr analogue TNFn-34 | | Descriptor: | 3-dodecylsulfanylpropanoic acid, NAD-dependent protein deacetylase sirtuin-2, Peptide-based TNFa-Myr analogue TNFn-34, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
6PVI
 
 | | Crystal structure of PhqK in complex with paraherquamide L | | Descriptor: | (8aS,13S,13aR,14aS)-4,4,13,15,15-pentamethyl-12,13,14,14a,15,16-hexahydro-4H,8H,9H,11H-8a,13a-(epiminomethano)[1,4]dioxepino[2,3-a]indolizino[6,7-h]carbazol-17-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVF
 
 | | Crystal structure of PhqK in complex with malbrancheamide B | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
5HLB
 
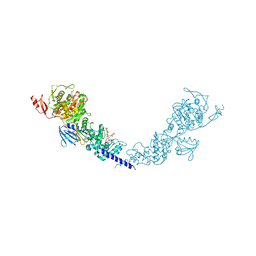 | | E. coli PBP1b in complex with acyl-aztreonam and moenomycin | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Escherichia coli Penicillin-Binding Protein 1B: Structural Insights into Inhibition.
J. Biol. Chem., 2016
|
|
6PVH
 
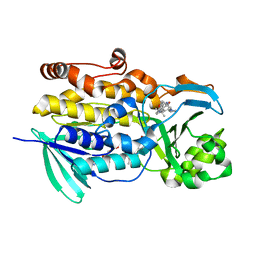 | | Crystal structure of PhqK in complex with paraherquamide K | | Descriptor: | (7aS,12S,12aR,13aS)-3,3,12,14,14-pentamethyl-3,7,11,12,13,13a,14,15-octahydro-8H,10H-7a,12a-(epiminomethano)indolizino[6,7-h]pyrano[3,2-a]carbazol-16-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
8ZTP
 
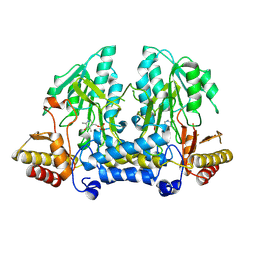 | | Crystal structure of cysteine desulfurase Sufs from Mycoplasma Pneumonia | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, cysteine desulfurase | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
7EK0
 
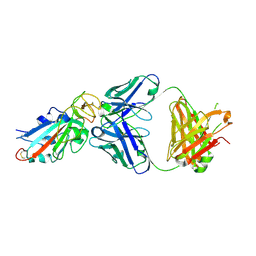 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7RUG
 
 | | Human SERINC3-DeltaICL4 | | Descriptor: | Serine incorporator 3, SiA | | Authors: | Purdy, M.D, Leonhardt, S.A, Yeager, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry.
Nat Commun, 14, 2023
|
|
8ZTQ
 
 | | Crystal structure of Sufu from Mycoplasma Pneumonia | | Descriptor: | Nitrogen fixation protein NifU, ZINC ION | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
5E1Y
 
 | | PDZ2 of LNX2 at 277K, model with alternate conformations | | Descriptor: | Ligand of Numb protein X 2 | | Authors: | Hekstra, D.R, White, K.I, Socolich, M.A, Ranganathan, R. | | Deposit date: | 2015-09-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.011 Å) | | Cite: | Electric-field-stimulated protein mechanics.
Nature, 540, 2016
|
|
8QT1
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-5 | | Descriptor: | (2S)-2-dodecylsulfanylpropanoic acid, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
