6FKI
 
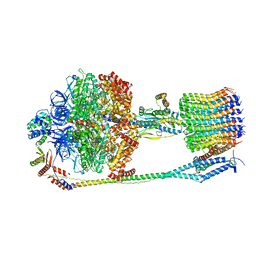 | | Chloroplast F1Fo conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
6HZZ
 
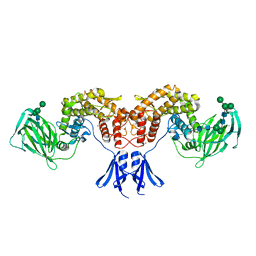 | | Structure of human D-glucuronyl C5 epimerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8P82
 
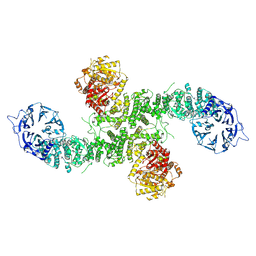 | | Cryo-EM structure of dimeric UBR5 | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
6Q7E
 
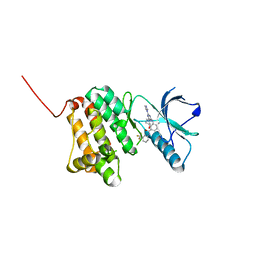 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATDL14 | | Descriptor: | 3-[[4-imidazol-1-yl-6-[(3~{R})-3-oxidanylpyrrolidin-1-yl]-1,3,5-triazin-2-yl]amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.059 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
5JBT
 
 | | Mesotrypsin in complex with cleaved amyloid precursor like protein 2 inhibitor (APLP2) | | Descriptor: | Amyloid-like protein 2, CALCIUM ION, PRSS3 protein, ... | | Authors: | Kayode, O, Wang, R, Pendlebury, D, Soares, A, Radisky, E.S. | | Deposit date: | 2016-04-13 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Acrobatic Substrate Metamorphosis Reveals a Requirement for Substrate Conformational Dynamics in Trypsin Proteolysis.
J. Biol. Chem., 291, 2016
|
|
7TY7
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Bicarbonate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY6
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatodihydrostilbene-2,2'-Disulfonic Acid (H2DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8P83
 
 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
7SVQ
 
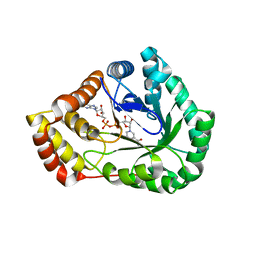 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea in complex with NAD+ | | Descriptor: | L-galactose dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
4WRR
 
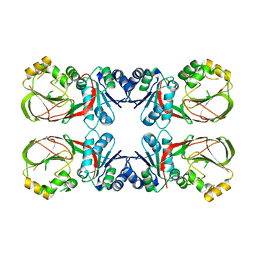 | | A putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | MAGNESIUM ION, putative diacylglycerol kinase | | Authors: | Hou, J, Zheng, H, Cooper, D.R, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
to be published
|
|
7TY4
 
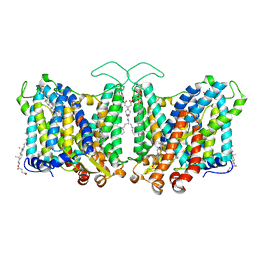 | | Cryo-EM structure of human Anion Exchanger 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5NED
 
 | |
7TYA
 
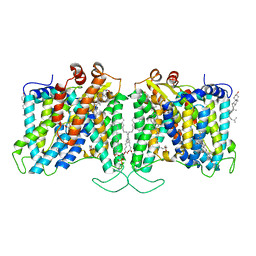 | | Cryo-EM structure of human Anion Exchanger 1 modified with Diethyl Pyrocarbonate (DEPC) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY8
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Niflumic Acid | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OWA
 
 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-4 | | Descriptor: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | Authors: | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
3ISB
 
 | | Binary complex of human DNA polymerase beta with a gapped DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Beard, W.A, Shock, D.D, Batra, V.K, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA polymerase beta substrate specificity: side chain modulation of the "A-rule".
J.Biol.Chem., 284, 2009
|
|
4WRT
 
 | | Crystal structure of Influenza B polymerase with bound vRNA promoter (form FluB2) | | Descriptor: | Influenza virus polymerase vRNA promoter 3' end, Influenza virus polymerase vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
7TPI
 
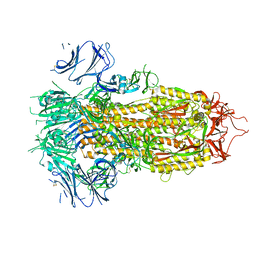 | | SARS-CoV-2 E406W mutant Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Addetia, A, Veesler, D. | | Deposit date: | 2022-01-25 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | SARS-CoV-2 E406W mutant Spike ectodomain
To Be Published
|
|
4WS9
 
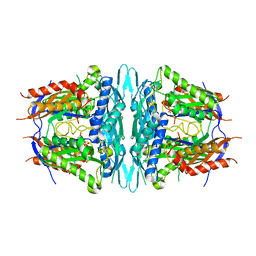 | | Crystal structure of sMAT N159G from Sulfolobus solfataricus | | Descriptor: | PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Wang, F, Brady, E.L, Singh, S, Clinger, J.A, Huber, T.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of sMAT N159G from Sulfolobus solfataricus.
To Be Published
|
|
5JMF
 
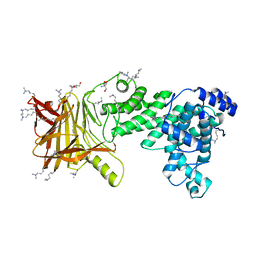 | | Heparinase III-BT4657 gene product | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Heparinase III protein, ... | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
7T12
 
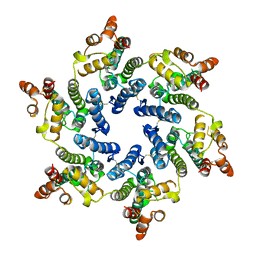 | | Hexameric HIV-1 (O-group) CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
8FIG
 
 | |
6W23
 
 | | ClpA Disengaged State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6NEG
 
 | |
4WZZ
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTAS (Cphy_0583, TARGET EFI-511148) WITH BOUND L-RHAMNOSE | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transporter, substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTAS (Cphy_0583, TARGET EFI-511148) WITH BOUND L-RHAMNOSE
To be published
|
|
