7RSF
 
 | | Acetylornithine deacetylase from Escherichia coli | | Descriptor: | Acetylornithine deacetylase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Endres, M, Becker, D.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acetylornithine deacetylase from Escherichia coli
To Be Published
|
|
5JGM
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with ethyl (2-(methylamino)-8-oxo-7,8-dihydro-1H-imidazo[4,5-g]quinazolin-6-yl)carbamate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Carbamate-protected lin-Benzoguanines as promising candidates for improving solubility and membrane permeability
To be Published
|
|
6OV7
 
 | | CFTR Associated Ligand (CAL) PDZ domain bound to peptide kCAL01 | | Descriptor: | Golgi-associated PDZ and coiled-coil motif-containing protein, kCAL01 peptide | | Authors: | Gill, N.P, Madden, D.R. | | Deposit date: | 2019-05-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Computational Analysis of Energy Landscapes Reveals Dynamic Features That Contribute to Binding of Inhibitors to CFTR-Associated Ligand.
J.Phys.Chem.B, 123, 2019
|
|
6O4I
 
 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6THE
 
 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6YNI
 
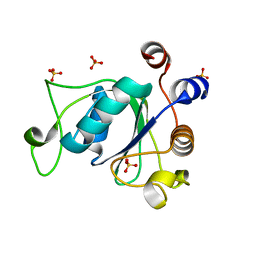 | | Crystal structure of YTHDC1 with compound DHU_DC1_036 | | Descriptor: | (2~{R},3~{R})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNL
 
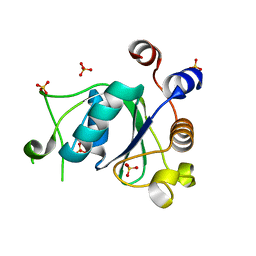 | | Crystal structure of YTHDC1 with compound DHU_DC1_078 | | Descriptor: | 3-[(2~{R})-5,5-dimethyl-2-phenyl-morpholin-4-yl]-~{N}-(methylcarbamoyl)propanamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6OD9
 
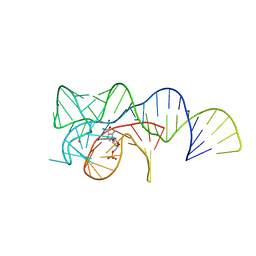 | | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CESIUM ION, MAGNESIUM ION, ... | | Authors: | Jones, C.P, Tran, B, Ferre-D'Amare, A.R. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.102 Å) | | Cite: | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5IZV
 
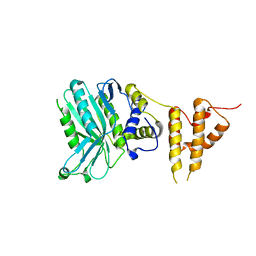 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
7S6O
 
 | | The crystal structure of Lys48-linked di-ubiquitin | | Descriptor: | ACETATE ION, Ubiquitin | | Authors: | Osipiuk, J, Tesar, C, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
6TUI
 
 | | Virion of empty GTA particle | | Descriptor: | Adaptor protein Rcc01688, Phage major capsid protein, HK97 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (10.47 Å) | | Cite: | Virion of empty GTA particle
Nat Commun, 11, 2020
|
|
8HHK
 
 | |
5I02
 
 | | tRNA guanine transglycosylase (TGT) in co-crystallized complex with 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 1,2-ETHANEDIOL, 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, ... | | Authors: | Ehrmann, F.R, Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Co-crystallization, Isothermal titration calorimetry and nanoESI-MS reveal dimer disturbing inhibitors
To be Published
|
|
3BJ5
 
 | | Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b' domain | | Descriptor: | Protein disulfide-isomerase, SULFATE ION | | Authors: | Ruddock, L.W, Nguyen, V.D, Wierenga, R.K, Haapalainen, A.M. | | Deposit date: | 2007-12-03 | | Release date: | 2008-09-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b' domain
J.Mol.Biol., 383, 2008
|
|
1DSM
 
 | | (-)-duocarmycin SA covalently linked to duplex DNA | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, 5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3' | | Authors: | Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structural basis for in situ activation of DNA alkylation by duocarmycin SA
J.Mol.Biol., 300, 2000
|
|
4J3J
 
 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
6YR8
 
 | | Affimer K6 - KRAS protein complex | | Descriptor: | Affimer K6, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
8HHI
 
 | |
7S7Y
 
 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein | | Descriptor: | IMIDAZOLE, iNicSnFR 4.0 Fluorescent Nicotine Sensor precursor binding protein | | Authors: | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
5I07
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with 6-Amino-4-(2-phenylethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINO-4-(2-PHENYLETHYL)-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Co-crystallization, Isothermal titration calorimetry and nanoESI-MS reveal dimer disturbing inhibitors
To be Published
|
|
5IAS
 
 | | Caspase 3 V266Y | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7RRM
 
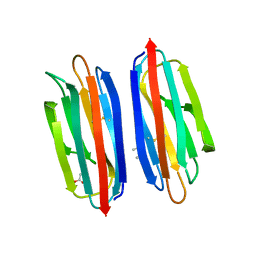 | | Structure of the human TMED1 (p24gamma1) Golgi dynamics Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Transmembrane emp24 domain-containing protein 1 | | Authors: | Mota, D.C.A.M, Cardoso, I.A, Mori, R.M, Mendes, L.F.S, Nonato, M.C, Filho, A.J.C. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and thermodynamic analyses of human TMED1 (p24 gamma 1) Golgi dynamics.
Biochimie, 192, 2022
|
|
7RWL
 
 | | Envelope-associated Adeno-associated virus serotype 2 | | Descriptor: | Capsid protein VP1 | | Authors: | Hull, J.A, Mietzsch, M, Chipman, P, Strugatsky, D, McKenna, R. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization of an envelope-associated adeno-associated virus type 2 capsid.
Virology, 565, 2021
|
|
7RWT
 
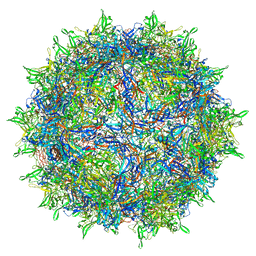 | | Adeno-associated virus type 2 | | Descriptor: | Capsid protein VP1 | | Authors: | Hull, J.A, Mietzsch, M, Chipman, P, Strugatsky, D, McKenna, R. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural characterization of an envelope-associated adeno-associated virus type 2 capsid.
Virology, 565, 2021
|
|
