6WZS
 
 | | Fusibacterium ulcerans ZTP riboswitch bound to m-1-pyridinyl AICA | | Descriptor: | 5-amino-1-(pyridin-3-yl)-1H-imidazole-4-carboxamide, Fusibacterium ulcerans ZTP riboswitch, MAGNESIUM ION, ... | | Authors: | Pichling, P, Jones, C.P, Ferre-D'Amare, A.R, Tran, B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design.
Cell Chem Biol, 27, 2020
|
|
6WZR
 
 | | Fusibacterium ulcerans ZTP riboswitch bound to p-1-pyridinyl AICA | | Descriptor: | 5-amino-1-(pyridin-4-yl)-1H-imidazole-4-carboxamide, Fusibacterium ulcerans ZTP riboswitch, MAGNESIUM ION, ... | | Authors: | Pichling, P, Jones, C.P, Ferre-D'Amare, A.R, Tran, B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design.
Cell Chem Biol, 27, 2020
|
|
6OD9
 
 | | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CESIUM ION, MAGNESIUM ION, ... | | Authors: | Jones, C.P, Tran, B, Ferre-D'Amare, A.R. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.102 Å) | | Cite: | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6OJN
 
 | | Comparative Model of SGIV Major Coat Protein (MCP) Trimer Based on Cryo-EM Map | | Descriptor: | Major capsid protein | | Authors: | Pintilie, G, Chen, D.-H, Tran, B.N, Jakana, J, Wu, J, Hew, C.L, Chiu, W. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Segmentation and Comparative Modeling in an 8.6- angstrom Cryo-EM Map of the Singapore Grouper Iridovirus.
Structure, 27, 2019
|
|
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
1RF3
 
 | | Structurally Distinct Recognition Motifs in Lymphotoxin-B Receptor and CD40 for TRAF-mediated Signaling | | Descriptor: | 24-residue peptide from Lymphotoxin-B Receptor, TNF receptor associated factor 3 | | Authors: | Li, C, Norris, P.S, Ni, C.Z, Havert, M.L, Chiong, E.M, Tran, B.R, Cabezas, E, Cheng, G, Reed, J.C, Satterthwait, A.C, Ware, C.F, Ely, K.R. | | Deposit date: | 2003-11-07 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structurally distinct recognition motifs in lymphotoxin-beta receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling.
J.Biol.Chem., 278, 2003
|
|
2N3Y
 
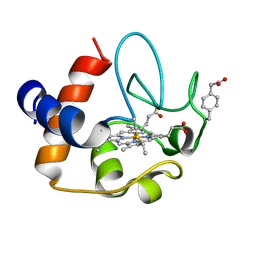 | | NMR structure of the Y48pCMF variant of human cytochrome c in its reduced state | | Descriptor: | Cytochrome c, Mesoheme | | Authors: | Moreno-Beltran, B, Del Conte, R, Diaz-Quintana, A, De la Rosa, M.A, Turano, P, Diaz-Moreno, I. | | Deposit date: | 2015-06-15 | | Release date: | 2016-12-14 | | Last modified: | 2017-04-26 | | Method: | SOLUTION NMR | | Cite: | Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7ZOL
 
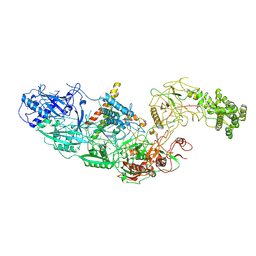 | | Cryo-EM structure of a CRISPR effector in complex with regulator | | Descriptor: | Cas7-11, TPR-CHAT, ZINC ION, ... | | Authors: | Babatunde, E.E, Davide, T, Bertrand, B, Sergey, N, Alexander, M, Henning, S, Dong, C.N. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the regulation of Cas7-11 by TPR-CHAT.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZOQ
 
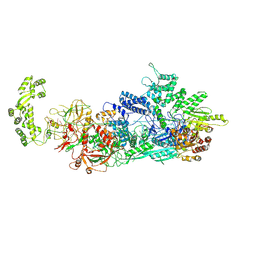 | | Cryo-EM structure of a CRISPR effector in complex with a caspase regulator | | Descriptor: | RNA (39-MER), TPR-CHAT, ZINC ION, ... | | Authors: | Babatunde, E.E, Davide, T, Bertrand, B, Sergey, N, Alexander, M, Henning, S, Dong, C.N. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the regulation of Cas7-11 by TPR-CHAT.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1HAR
 
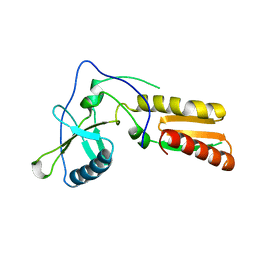 | |
