8F3M
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant with S466 insertion apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3G
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant in the penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3Q
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) Y460A variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
6YRI
 
 | |
6ZR9
 
 | |
8CK0
 
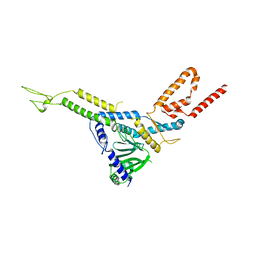 | |
7RCB
 
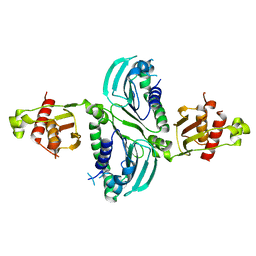 | | Crystal Structure of a PMS2 VUS | | Descriptor: | Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7RCI
 
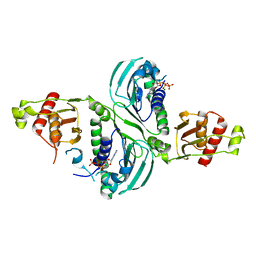 | | Crystal Structure of a PMS2 VUS with Substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7RCK
 
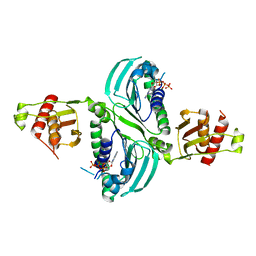 | | Crystal Structure of PMS2 with Substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
8F43
 
 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
6FTK
 
 | | Gp36-MPER | | Descriptor: | Envelope protein | | Authors: | D'Ursi, A.M, Grimaldi, M. | | Deposit date: | 2018-02-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the FIV gp36 C-Terminal Heptad Repeat and Membrane-Proximal External Region.
Int J Mol Sci, 21, 2020
|
|
7JYD
 
 | | Human Liver Receptor Homolog-1 in Complex with 10CA and a Fragment of Tif2 | | Descriptor: | 10-[(3aR,6R,6aR)-6-hydroxy-3-phenyl-3a-(1-phenylethenyl)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
7JYE
 
 | | Human Liver Receptor Homolog-1 in Complex with 9ChoP and a Fragment of Tif2 | | Descriptor: | 9-[(3~{a}~{R},6~{R},6~{a}~{R})-6-oxidanyl-3-phenyl-3~{a}-(1-phenylethenyl)-4,5,6,6~{a}-tetrahydro-1~{H}-pentalen-2-yl]nonyl 2-(trimethyl-$l^{4}-azanyl)ethyl hydrogen phosphate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
8SP5
 
 | |
8D35
 
 | |
5BRD
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor BENZ-GlcN | | Descriptor: | 2-(benzoylamino)-2-deoxy-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
5BRF
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor HPOP-GlcN | | Descriptor: | 2-deoxy-2-{[3-(4-hydroxyphenyl)propanoyl]amino}-alpha-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
5BRE
 
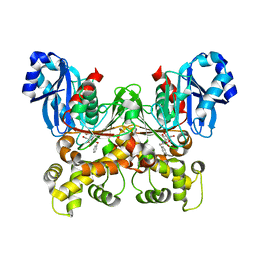 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN | | Descriptor: | 2-{[(benzyloxy)carbonyl]amino}-2-deoxy-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
5WG8
 
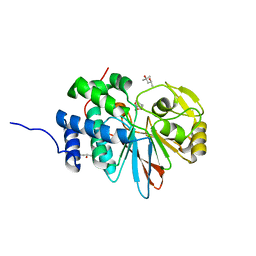 | | Structure of PP5C with LB-100; 7-oxabicyclo[2.2.1]heptane-2,3-dicarbonyl moiety modeled in the density | | Descriptor: | (1S,2R,3S,4R)-3-(4-methylpiperazine-1-carbonyl)-7-oxabicyclo[2.2.1]heptane-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | D'Arcy, B.M, Swingle, M.R, Honkanen, R.E, Prakash, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Antitumor Drug LB-100 Is a Catalytic Inhibitor of Protein Phosphatase 2A (PPP2CA) and 5 (PPP5C) Coordinating with the Active-Site Catalytic Metals in PPP5C.
Mol. Cancer Ther., 18, 2019
|
|
5BRH
 
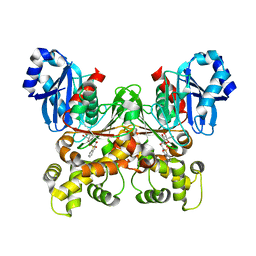 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor DBT-GlcN | | Descriptor: | 2-deoxy-2-({[(1,1-dioxido-1-benzothiophen-2-yl)methoxy]carbonyl}amino)-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
4IU5
 
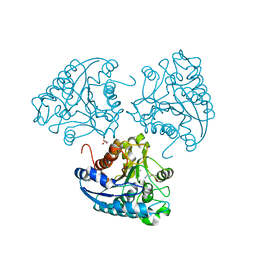 | | Crystal structure of Leishmania mexicana arginase in complex with catalytic product L-ornithine | | Descriptor: | Arginase, GLYCEROL, L-ornithine, ... | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
4IU0
 
 | | Crystal structure of Leishmania mexicana arginase in complex with inhibitor ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, GLYCEROL, ... | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
4ITY
 
 | | Crystal structure of Leishmania mexicana arginase | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
4IU4
 
 | | Crystal structure of Leishmania mexicana arginase in complex with inhibitor BEC | | Descriptor: | Arginase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
4IU1
 
 | | Crystal structure of Leishmania mexicana arginase in complex with inhibitor nor-NOHA | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
