5DLC
 
 | |
7UJ8
 
 | |
6MRK
 
 | |
4Z6P
 
 | | Structure of H200Q variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with HPCA at 1.75 Ang resolution | | 分子名称: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, CHLORIDE ION, FE (II) ION, ... | | 著者 | Kovaleva, E.G, Lipscomb, J.D. | | 登録日 | 2015-04-06 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
6TKZ
 
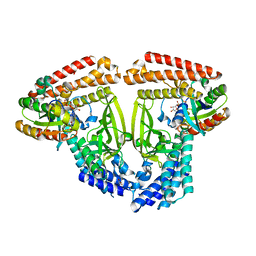 | |
5JG6
 
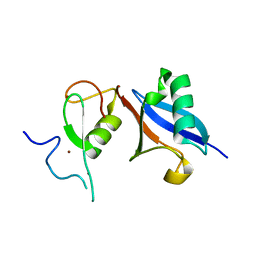 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | 分子名称: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | 著者 | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | 登録日 | 2016-04-19 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.0013 Å) | | 主引用文献 | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
6WNX
 
 | | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide | | 分子名称: | Catenin beta-1, F-box/WD repeat-containing protein 11, GLYCEROL, ... | | 著者 | Ivanochko, D, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Boettcher, J, Structural Genomics Consortium (SGC) | | 登録日 | 2020-04-23 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide
To Be Published
|
|
6QZ8
 
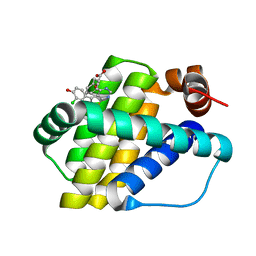 | | Structure of Mcl-1 in complex with compound 10d | | 分子名称: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6GRI
 
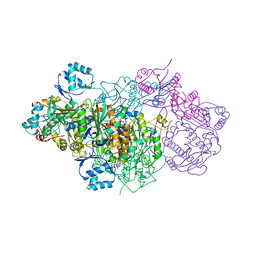 | | E. coli Microcin synthetase McbBCD complex | | 分子名称: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Microcin B17-processing protein McbB, ... | | 著者 | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | 登録日 | 2018-06-11 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
3JRE
 
 | |
6QU9
 
 | | Fab fragment of an antibody that inhibits polymerisation of alpha-1-antitrypsin | | 分子名称: | FAB 4B12 heavy chain, FAB 4B12 light chain, GLYCEROL, ... | | 著者 | Jagger, A.M, Heyer-Chauhan, N, Lomas, D.A, Irving, J.A. | | 登録日 | 2019-02-26 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structural basis for Z alpha 1 -antitrypsin polymerization in the liver.
Sci Adv, 6, 2020
|
|
5E9Z
 
 | | Cytochrome P450 BM3 mutant M11 | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, FE (II) ION, ... | | 著者 | Capoferri, L, Leth, R, ter Haar, E, Mohanty, A.K, Grootenhuis, D.J, Vottero, E, Commandeur, J.N.M, Vermeulen, N.P.E, Jorgensen, F.S, Olsen, L, Geerke, D.P. | | 登録日 | 2015-10-15 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Insights into regioselective metabolism of mefenamic acid by cytochrome P450 BM3 mutants through crystallography, docking, molecular dynamics, and free energy calculations.
Proteins, 84, 2016
|
|
8AWS
 
 | | Millisecond cryo-trapping by the spitrobot crystal plunger, Xylose Isomerase with Glucose at 50ms | | 分子名称: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | 著者 | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Doepke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | 登録日 | 2022-08-30 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
6GVW
 
 | | Crystal structure of the BRCA1-A complex | | 分子名称: | BRCA1-A complex subunit Abraxas 1, BRCA1-A complex subunit RAP80, BRISC and BRCA1-A complex member 1, ... | | 著者 | Bunker, R.D, Rabl, J, Thoma, N.H. | | 登録日 | 2018-06-21 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.75 Å) | | 主引用文献 | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
5NKT
 
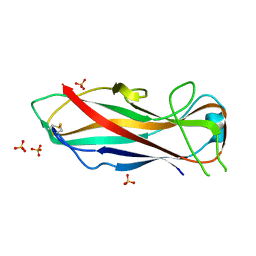 | | FimA wt from E. coli | | 分子名称: | SULFATE ION, Type-1 fimbrial protein, A chain | | 著者 | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | 登録日 | 2017-04-03 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
5NKU
 
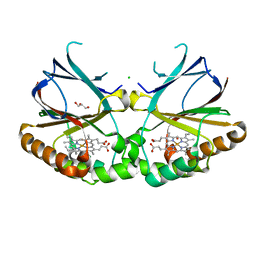 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2017-04-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
6QYP
 
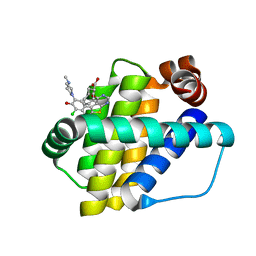 | | Structure of Mcl-1 in complex with compound 13 | | 分子名称: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-09 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ6
 
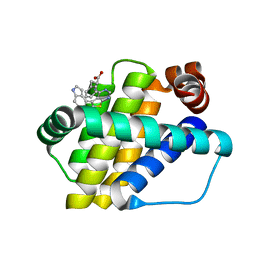 | | Structure of Mcl-1 in complex with compound 8b | | 分子名称: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
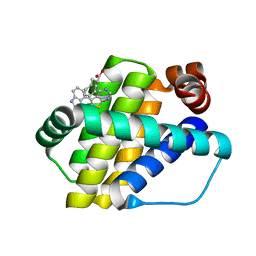 | | Structure of Mcl-1 in complex with compound 8a | | 分子名称: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
7CCR
 
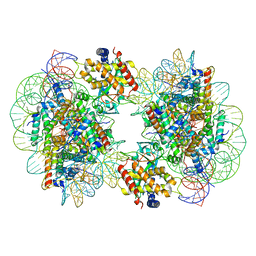 | | Structure of the 2:2 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
5WI8
 
 | |
6TW3
 
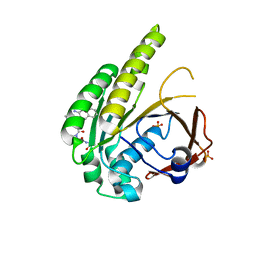 | | HumRadA2 in complex with Naphthyl-HPA fragment-peptide chimera | | 分子名称: | (2~{S})-1-[(2~{S})-2-[(3-azanylnaphthalen-2-yl)carbonylamino]-3-(1~{H}-imidazol-4-yl)propanoyl]-~{N}-[(2~{S})-1-azanyl-1-oxidanylidene-propan-2-yl]pyrrolidine-2-carboxamide, DNA repair and recombination protein RadA, PHOSPHATE ION | | 著者 | Marsh, M.E, Fischer, G, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | 登録日 | 2020-01-12 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.352 Å) | | 主引用文献 | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
7MKM
 
 | |
6QZ4
 
 | | Structure of MHETase from Ideonella sakaiensis | | 分子名称: | CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, SULFATE ION | | 著者 | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | 登録日 | 2019-03-11 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
