7NZM
 
 | | Cryo-EM structure of pre-dephosphorylation complex of phosphorylated eIF2alpha with trapped holophosphatase (PP1A_D64A/PPP1R15A/G-actin/DNase I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yan, Y, Hardwick, S, Ron, D. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OJY
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 6 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-oxidanyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OK1
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | Descriptor: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJ6
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, ACETATE ION, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJQ
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 7 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-methyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJW
 
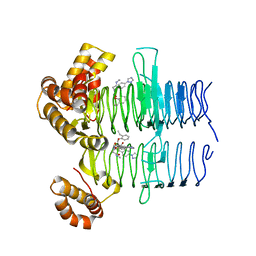 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 93 | | Descriptor: | 2-[2-(2-ethylphenoxy)ethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-~{N}-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OCV
 
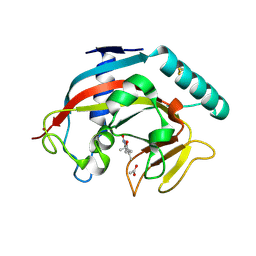 | | Human TNKS1 in complex with 3-[4-(1-Hydroxy-1-methyl-ethyl)-phenyl]-6-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one | | Descriptor: | 6-methyl-3-[4-(2-oxidanylpropan-2-yl)phenyl]-4~{H}-pyrrolo[1,2-a]pyrazin-1-one, ACETATE ION, Poly [ADP-ribose] polymerase, ... | | Authors: | Musil, D, Lehmann, M, Buchstaller, H.-P. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models.
J.Med.Chem., 64, 2021
|
|
7OK0
 
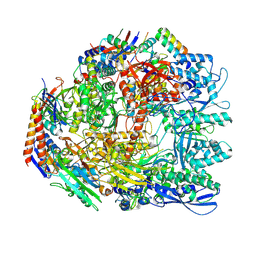 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7O82
 
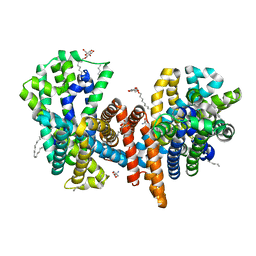 | | The L-arginine/agmatine antiporter from E. coli at 1.7 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine/agmatine antiporter, DECANE, ... | | Authors: | Jeckelmann, J.M, Ilgue, H, Kalbermatter, D, Fotiadis, D. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structure of the amino acid transporter AdiC reveals insights into the role of water molecules and networks in oligomerization and substrate binding.
Bmc Biol., 19, 2021
|
|
6CBJ
 
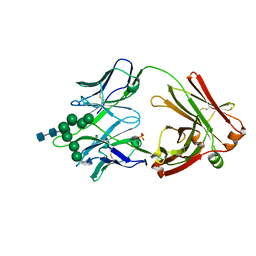 | | Crystal Structure of DH270.3 Fab in complex with Man9 | | Descriptor: | DH270.3 Fab heavy chain, DH270.3 Fab light chain, PHOSPHATE ION, ... | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope.
Nat Commun, 9, 2018
|
|
6CJ6
 
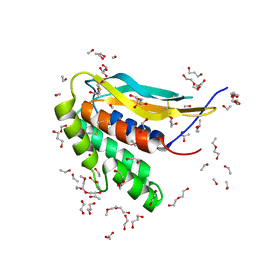 | | Structure of the poxvirus protein F9 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Diesterbeck, U.S, Gittis, A.G, Garboczi, D.N, Moss, B. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 angstrom structure of protein F9 and its comparison to L1, two components of the conserved poxvirus entry-fusion complex.
Sci Rep, 8, 2018
|
|
6CJX
 
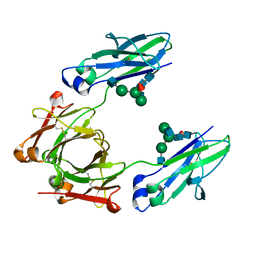 | | Crystal structure of a Fc fragment LALA mutant (L234A, L235A) of human IgG1 (crystal form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of human IgG1 | | Authors: | Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-02-27 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Antigen-Induced Allosteric Changes in a Human IgG1 Fc Increase Low-Affinity Fc gamma Receptor Binding.
Structure, 2020
|
|
6CXF
 
 | |
6CMG
 
 | |
5V7J
 
 | | Crystal Structure at 3.7 A Resolution of Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer with Four Glycans (N197, N276, N362, and N462) removed in Complex with Neutralizing Antibodies 3H+109L and 35O22. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 35O22 Fab heavy chain, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Kwong, P.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation.
Cell Rep, 19, 2017
|
|
6CMX
 
 | | Human Teneurin 2 extra-cellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, alpha-D-mannopyranose, ... | | Authors: | Shalev-Benami, M, Li, J, Sudhof, T, Skiniotis, G, Arac, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Cell, 173, 2018
|
|
7NWK
 
 | | Crystal structure of CDK9-Cyclin T1 bound by compound 6 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, N-((1R,3R)-3-(7-(4-fluoro-2-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-2-yl)cyclopentyl)acetamide | | Authors: | Collie, G.W, Ferguson, A.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
J.Med.Chem., 64, 2021
|
|
5UTF
 
 | | Crystal Structure of a Stabilized DS-SOSIP.6mut BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, Y177W, N300M, N302M, T320L, I420M in Complex with Human Antibodies PGT122 and 35O22 at 4.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35022 Heavy chain, ... | | Authors: | Pancera, M, Chuang, G.-Y, Xu, K, Kwong, P.D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
6CWE
 
 | | Structure of alpha-GSA[8,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(6-phenylhexyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycolipid recognition by iNKT cells
To Be Published
|
|
6CQV
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with VX(+) and HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
7NUW
 
 | | Crystal structure of the TIR domain of human TLR1 (crystallised with Zn2+ ions) | | Descriptor: | Toll-like receptor 1 | | Authors: | Vakhrameev, D.D, Luginina, A.P, Shevtsov, M.B, Lushpa, V.A, Mineev, K.S, Borshchevskiy, V.I. | | Deposit date: | 2021-03-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn 2+ ions.
Commun Biol, 4, 2021
|
|
7NUX
 
 | | Crystal structure of the TIR domain of human TLR1 (crystallised without ZN2+ ions) | | Descriptor: | Toll-like receptor 1 | | Authors: | Vakhrameev, D.D, Luginina, A.P, Shevtsov, M.B, Lushpa, V.A, Mineev, K.S, Borshchevskiy, V.I. | | Deposit date: | 2021-03-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn 2+ ions.
Commun Biol, 4, 2021
|
|
7ODY
 
 | |
6CQU
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase with Reactivator HI-6 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
5V6X
 
 | |
