6G68
 
 | |
7BJH
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 8 | | Descriptor: | CHLORIDE ION, N,N-dimethyl-7H-purin-6-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
6RRX
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-3-piperidinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)piperidin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
4YB0
 
 | | 3',3'-cGAMP riboswitch bound with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
6N21
 
 | | Structure of wild-type CAO1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
4YBB
 
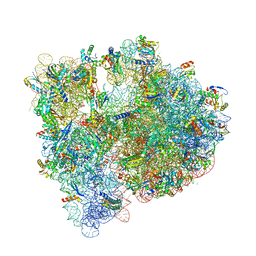 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Noeske, J, Wasserman, M.R, Terry, D.S, Altman, R.B, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structure of the Escherichia coli ribosome.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8U62
 
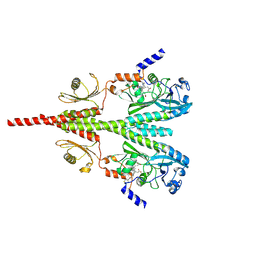 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers FL | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
7BJK
 
 | |
8QX8
 
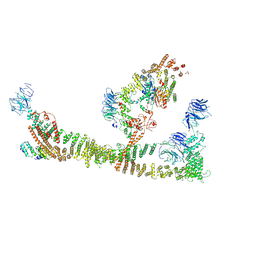 | | Endosomal membrane tethering complex CORVET | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar protein sorting-associated protein 16, ... | | Authors: | Shvarev, D, Ungermann, C, Moeller, A. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the endosomal CORVET tethering complex.
Nat Commun, 15, 2024
|
|
7B9S
 
 | | Structure of the mycobacterial ESX-5 Type VII Secretion System hexameric pore complex | | Descriptor: | EccB5, EccC5, EccD5, ... | | Authors: | Chojnowski, G, Ritter, C, Beckham, K.S.H, Mullapudi, E, Rettel, M, Savitski, M.M, Mortensen, S.A, Ziemianowicz, D, Kosinski, J, Wilmanns, M. | | Deposit date: | 2020-12-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mycobacterial ESX-5 type VII secretion system pore complex.
Sci Adv, 7, 2021
|
|
6GB8
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: small cell polymorph dataset 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7BKN
 
 | | Crystal structure of CHK1 complex with adenine | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Baker, L.M, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJE
 
 | | Crystal structure of CHK1-10pt-mutant complex with adenine | | Descriptor: | ADENINE, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
4YMB
 
 | | Structure of the ligand-binding domain of GluK1 in complex with the antagonist CNG10111 | | Descriptor: | (3R,4S)-3-(3-carboxyphenyl)-4-propyl-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
6RUT
 
 | |
6QLR
 
 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative 5 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
8ACA
 
 | | SDBC DR_0644 subunit, only-Cu Superoxide Dismutase | | Descriptor: | COPPER (II) ION, DR_0644, only-Cu Superoxide Dismutase | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
4Z2F
 
 | | Sclerotium Rolfsii lectin variant (SSR2) with fine carbohydrate specificity | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, lectin variant 2 | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
6TA7
 
 | | CRYSTAL STRUCTURE OF HUMAN G3BP1-NTF2 IN COMPLEX WITH HUMAN CAPRIN1-DERIVED SOLOMON MOTIF | | Descriptor: | CHLORIDE ION, Caprin-1, Ras GTPase-activating protein-binding protein 1, ... | | Authors: | Schulte, T, Achour, A, Panas, M.D, McInerney, G.M. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Caprin-1 binding to the critical stress granule protein G3BP1 is regulated by pH
Biorxiv, 2021
|
|
6QN9
 
 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
7BO7
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJB44355437 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[(4~{a}~{S},7~{a}~{S})-4-azanyl-1,4,4~{a},7~{a}-tetrahydropyrrolo[2,3-d]pyrimidin-7-yl]-5-(quinolin-7-yloxymethyl)oxolane-3,4-diol, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Brown, D, Robinson, C, Carr, K.H, Pande, V. | | Deposit date: | 2021-01-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJB44355437
To Be Published
|
|
8EBN
 
 | | Structure of KLHDC2-EloB/C tetrameric assembly | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 2 | | Authors: | Scott, D.C, Schulman, B.A. | | Deposit date: | 2022-08-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E3 ligase autoinhibition by C-degron mimicry maintains C-degron substrate fidelity.
Mol.Cell, 83, 2023
|
|
6FR2
 
 | | Soluble epoxide hydrolase in complex with LK864 | | Descriptor: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
1CZQ
 
 | | CRYSTAL STRUCTURE OF THE D10-P1/IQN17 COMPLEX: A D-PEPTIDE INHIBITOR OF HIV-1 ENTRY BOUND TO THE GP41 COILED-COIL POCKET. | | Descriptor: | CHLORIDE ION, D-PEPTIDE INHIBITOR, FUSION PROTEIN BETWEEN THE HYDROPHOBIC POCKET OF HIV GP41 AND GCN4-PIQI | | Authors: | Eckert, D.M, Malashkevich, V.N, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 1999-09-06 | | Release date: | 1999-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket.
Cell(Cambridge,Mass.), 99, 1999
|
|
6N1Y
 
 | | Structure of L509V CAO1 - growth condition 1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
