4Y1D
 
 | |
7AZ5
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
6UZN
 
 | |
6UZS
 
 | |
5MQN
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | CALCIUM ION, Glycosyl hydrolases family 2, sugar binding domain | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
7O2R
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ITF3985 | | Descriptor: | 3,5-bis(fluoranyl)-~{N}-oxidanyl-4-[(5-pyrimidin-2-yl-1,2,3,4-tetrazol-2-yl)methyl]benzamide, DI(HYDROXYETHYL)ETHER, Histone deacetylase 6, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
5MU2
 
 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, synthetic peptide containing the CII583-591 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
7AZE
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
6SS4
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
8HKQ
 
 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8C1L
 
 | | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Hepatocyte nuclear factor 4-alpha, Nuclear receptor coactivator 2, ... | | Authors: | Ni, X, Merk, D, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide
To Be Published
|
|
8HKK
 
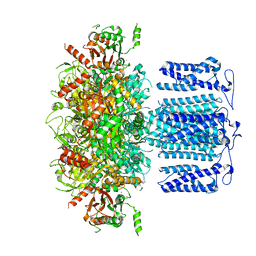 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
6Y6B
 
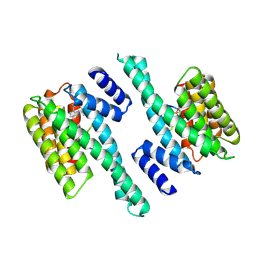 | | Crystal structure of human 14-3-3 gamma in complex with CaMKK2 14-3-3 binding motif Ser100 and 16-OMe-Fusicoccin H | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[[(1~{S},3~{R},6~{S},7~{S},8~{R},9~{R},10~{R},11~{R},14~{S})-14-(me thoxymethyl)-3,10-dimethyl-9-oxidanyl-6-propan-2-yl-8-tricyclo[9.3.0.0^{3,7}]tetradecanyl]oxy]oxane-3,4,5-triol, 14-3-3 protein gamma, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Lentini Santo, D, Obsilova, V, Obsil, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization of Protein-Protein Interactions between CaMKK2 and 14-3-3 by Fusicoccins.
Acs Chem.Biol., 15, 2020
|
|
7AZK
 
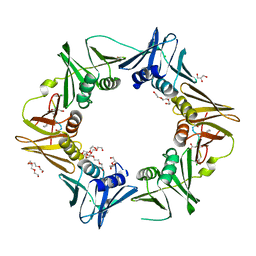 | | DNA polymerase sliding clamp from Escherichia coli with peptide 35 bound | | Descriptor: | Beta sliding clamp, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
6MVO
 
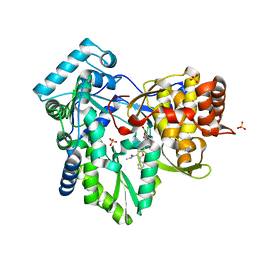 | | HCV NS5B 1A Y316 bound to Compound 49 | | Descriptor: | 6-[(7-chloro-1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)(methylsulfonyl)amino]-5-cyclopropyl-2-(4-fluorophenyl)-N-methyl-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
8HIR
 
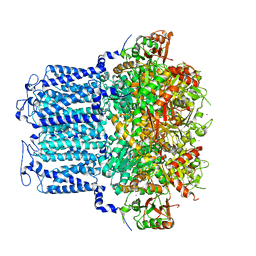 | | potassium channels | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
3ILR
 
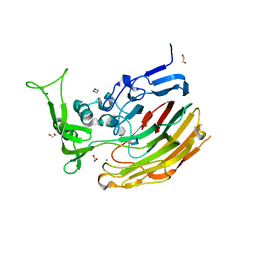 | | Structure of Heparinase I from Bacteroides thetaiotaomicron in complex with tetrasaccharide product | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(4-1)-4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid, 4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | Authors: | Garron, M.L, Cygler, M, Shaya, D. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
8HKM
 
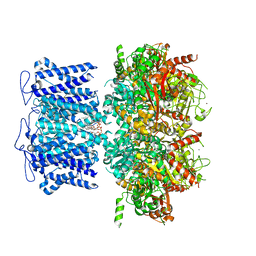 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HK6
 
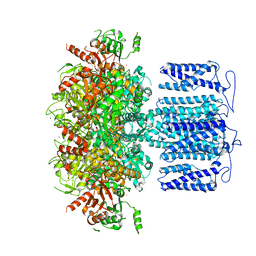 | | potassium channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
6Y8S
 
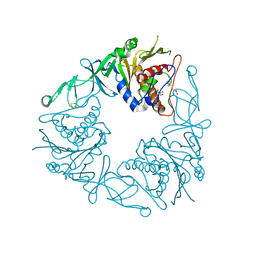 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6MVP
 
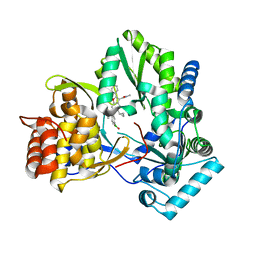 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}phenyl)boronic acid, Genome polyprotein | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
7TFS
 
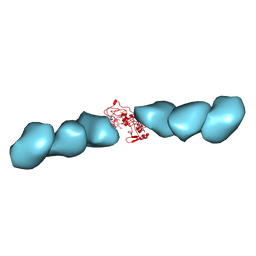 | | Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-04 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
8HKF
 
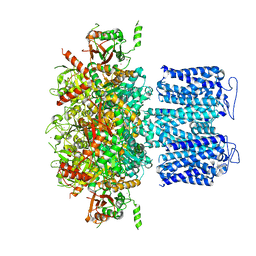 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, Z.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
6BHY
 
 | | Mouse Immunoglobulin G 2c Fc fragment with single GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, SODIUM ION | | Authors: | Falconer, D, Barb, A. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mouse IgG2c Fc loop residues promote greater receptor-binding affinity than mouse IgG2b or human IgG1.
PLoS ONE, 13, 2018
|
|
