1Y10
 
 | | Mycobacterial adenylyl cyclase Rv1264, holoenzyme, inhibited state | | Descriptor: | CALCIUM ION, Hypothetical protein Rv1264/MT1302, PENTAETHYLENE GLYCOL | | Authors: | Tews, I, Findeisen, F, Sinning, I, Schultz, A, Schultz, J.E, Linder, J.U. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme
Science, 308, 2005
|
|
3BTJ
 
 | |
3BTL
 
 | |
1OLS
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
6ETJ
 
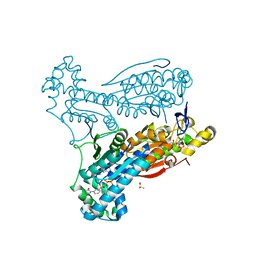 | | HUMAN PFKFB3 IN COMPLEX WITH KAN0438241 | | Descriptor: | 4-[[3-(5-fluoranyl-2-oxidanyl-phenyl)phenyl]sulfonylamino]-2-oxidanyl-benzoic acid, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Gustafsson, N.M.S, Lundback, T, Farnegardh, K, Groth, P, Wiitta, E, Jonsson, M, Hallberg, K, Pennisi, R, Huguet Ninou, A, Martinsson, J, Norstrom, C, Schultz, J, Andersson, M, Markova, N, Marttila, P, Norin, M, Olin, T, Helleday, T. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination.
Nat Commun, 9, 2018
|
|
3BTI
 
 | |
1ZMD
 
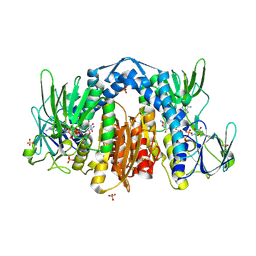 | | Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Human Dihydrolipoamide Dehydrogenase: NAD+/NADH Binding and the Structural Basis of Disease-causing Mutations
J.Mol.Biol., 350, 2005
|
|
3VAD
 
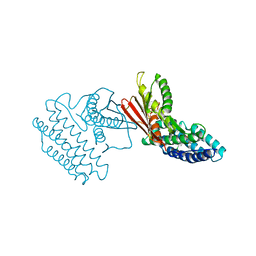 | | Crystal structure of I170F mutant branched-chain alpha-ketoacid dehydrogenase kinase in complex with 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ahmed, K, Gui, W.J, Tso, S.C, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2011-12-29 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal structure of I170F mutant branched-chain alpha-ketoacid dehydrogenase kinase in complex with 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid
To be Published
|
|
3EXE
 
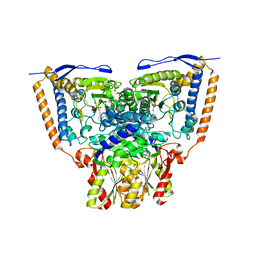 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3RNM
 
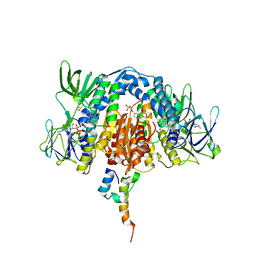 | | The crystal structure of the subunit binding of human dihydrolipoamide transacylase (E2b) bound to human dihydrolipoamide dehydrogenase (E3) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-MERCAPTOETHANOL, Dihydrolipoyl dehydrogenase, ... | | Authors: | Brautigam, C.A, Wynn, R.M, Chuang, J.C, Young, B.B, Chuang, D.T. | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Basis for Weak Interactions between Dihydrolipoamide Dehydrogenase and Subunit-binding Domain of the Branched-chain {alpha}-Ketoacid Dehydrogenase Complex.
J.Biol.Chem., 286, 2011
|
|
8V3S
 
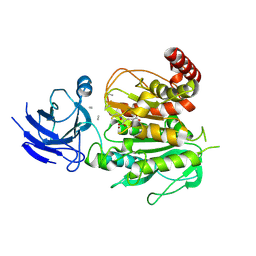 | | Structure of CCP5 class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3N
 
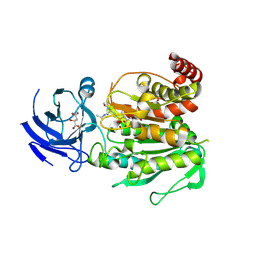 | | CCP5 in complex with Glu-P-Glu transition state analog | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3O
 
 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3P
 
 | | CCP5 in complex with Glu-P-peptide 2 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, Tubulin beta-2A chain, ZINC ION | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3M
 
 | | CCP5 apo structure | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3Q
 
 | | Structure of CCP5 class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
8V3R
 
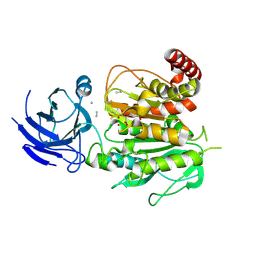 | | Structure of CCP5 class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation
Nature, 2024
|
|
7Q4V
 
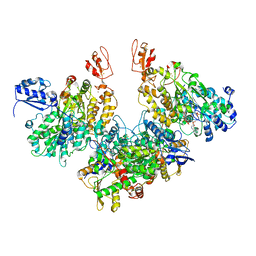 | | Electron bifurcating hydrogenase - HydABC from A. woodii | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Katsyv, A, Kumar, A, Saura, P, Poeverlein, M.C, Freibert, S.A, Stripp, S, Jain, S, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
7Q4W
 
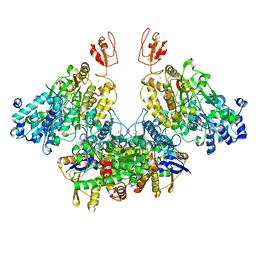 | | CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Poeverlein, M.C, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
8OH9
 
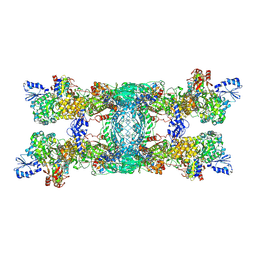 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH5
 
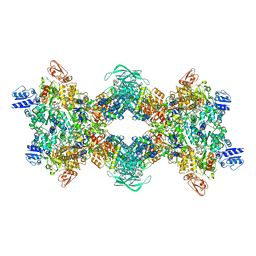 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
2Q8H
 
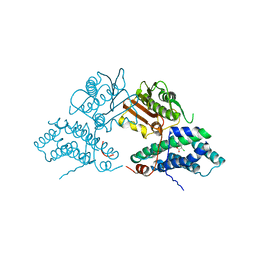 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with dichloroacetate (DCA) | | Descriptor: | DICHLORO-ACETIC ACID, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2Q8F
 
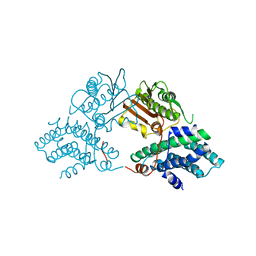 | | Structure of pyruvate dehydrogenase kinase isoform 1 | | Descriptor: | POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2II5
 
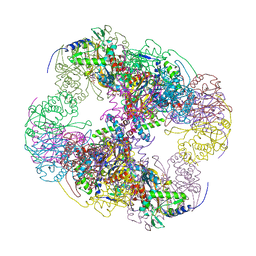 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Isobutyryl-Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, ISOBUTYRYL-COENZYME A, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2IHW
 
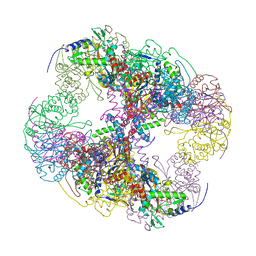 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), apo form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
