3G96
 
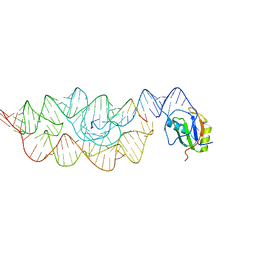 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
4RKQ
 
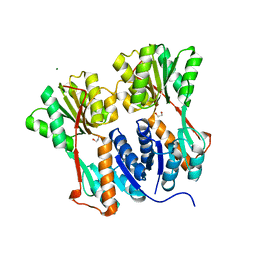 | | Crystal structure of LacI family transcriptional regulator from Arthrobacter sp. FB24, target EFI-560007 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator PurR from Arthrobacter Sp, Target Nysgxrc 11027R
To be Published
|
|
4RY0
 
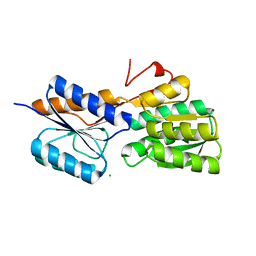 | | Crystal structure of ribose transporter solute binding protein RHE_PF00037 from Rhizobium etli CFN 42, TARGET EFI-511357, in complex with D-ribose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Probable ribose ABC transporter, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Ribose Transporter Solute Binding Protein from Rhizobium Etly, Target Efi-511357
To be Published
|
|
5LAM
 
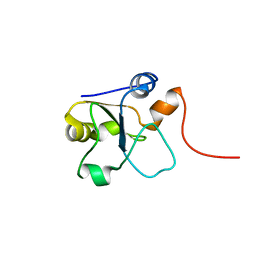 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
6II0
 
 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O | | Descriptor: | GLYCEROL, Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4RKR
 
 | | Crystal structure of LacI family transcriptional regulator from Arthrobacter sp. FB24, target EFI-560007, complex with lactose | | Descriptor: | Transcriptional regulator, LacI family, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator PurR from Arthrobacter Sp, Target Nysgxrc 11027R
To be Published
|
|
4RXT
 
 | | Crystal structure of carbohydrate transporter solute binding protein Arad_9553 from Agrobacterium Radiobacter, Target EFI-511541, in complex with D-arabinose | | Descriptor: | Sugar ABC transporter, alpha-L-arabinopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Sugar Binding Transporter Arad_9553 from Agrobacterium Radiobacter, Target Efi-511541
To be Published
|
|
4RS3
 
 | | Crystal structure of carbohydrate transporter A0QYB3 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with xylitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
6IM1
 
 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), TETRAETHYLENE GLYCOL, ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
4RXM
 
 | | Crystal structure of periplasmic ABC transporter solute binding protein A7JW62 from Mannheimia haemolytica PHL213, Target EFI-511105, in complex with Myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of solute binding sugar transporter A7JW62 from Mannheimia haemolytica, Target EFI-511105.
To be Published
|
|
3G8S
 
 | | Crystal structure of the pre-cleaved Bacillus anthracis glmS ribozyme | | Descriptor: | GLMS RIBOZYME, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
6II2
 
 | | Crystal structure of alpha-beta hydrolase (ABH) and Makes Caterpillars Floppy (MCF)-Like effectors of Vibrio vulnificus MO6-24/O | | Descriptor: | Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyung, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IM0
 
 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | BICARBONATE ION, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
5FKX
 
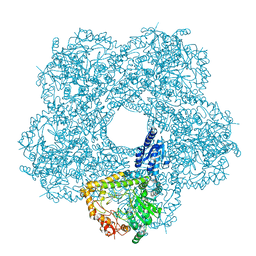 | | Structure of E.coli inducible lysine decarboxylase at active pH | | Descriptor: | LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
5FL2
 
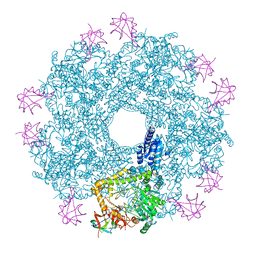 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
1YGM
 
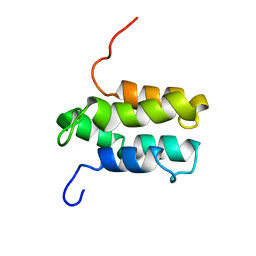 | | NMR structure of Mistic | | Descriptor: | hypothetical protein BSU31320 | | Authors: | Roosild, T.P, Greenwald, J, Vega, M, Castronovo, S, Riek, R, Choe, S. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mistic, a membrane-integrating protein for membrane protein expression.
Science, 307, 2005
|
|
1UOM
 
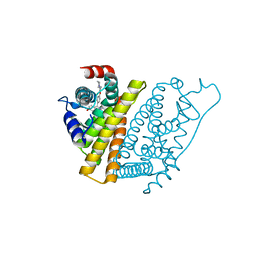 | | The Structure of Estrogen Receptor in Complex with a Selective and Potent Tetrahydroisochiolin Ligand. | | Descriptor: | 2-PHENYL-1-[4-(2-PIPERIDIN-1-YL-ETHOXY)-PHENYL]-1,2,3,4-TETRAHYDRO-ISOQUINOLIN-6-OL, ESTROGEN RECEPTOR | | Authors: | Stark, W, Bischoff, S.F, Buhl, T, Fournier, B, Halleux, C, Kallen, J, Keller, H, Renaud, J. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Estrogen Receptor Modulators: Identification and Structure-Activity Relationships of Potent Eralpha-Selective Tetrahydroisoquinoline Ligands
J.Med.Chem., 46, 2003
|
|
5HBP
 
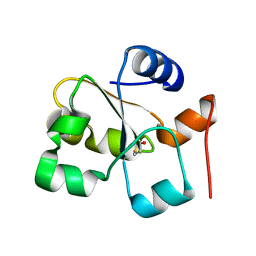 | | The crystal of rhodanese domain of YgaP treated with SNOC | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5FKZ
 
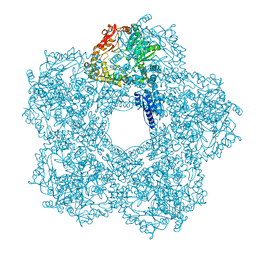 | | Structure of E.coli Constitutive lysine decarboxylase | | Descriptor: | LYSINE DECARBOXYLASE, CONSTITUTIVE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
4RU1
 
 | | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus 11B, TARGET EFI-510965, in complex with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CITRIC ACID, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus, TARGET EFI-510965.
To be Published
|
|
5LAO
 
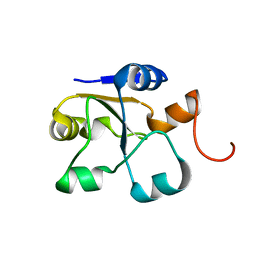 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
4RY8
 
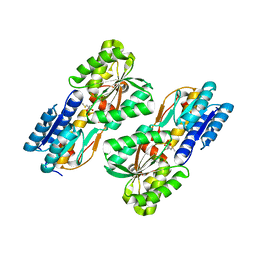 | | Crystal structure of 5-methylthioribose transporter solute binding protein TLET_1677 from Thermotoga lettingae TMO TARGET EFI-511109 in complex with 5-methylthioribose | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, Periplasmic binding protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of 5-methylthioribose binding protein TLET_1677 from Thermotoga lettingae TARGET EFI-511109
To be Published
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
