6F3K
 
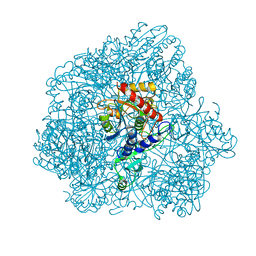 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6Z8K
 
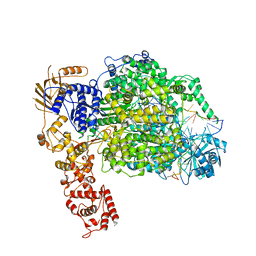 | | La Crosse virus polymerase at elongation mimicking stage | | Descriptor: | La Crosse virus 3' vRNA (1-16), La Crosse virus 5' vRNA (9-16), La Crosse virus 5' vRNA 1-10, ... | | Authors: | Arragain, B, Effantin, G, Schoehn, G, Cusack, S, Malet, H. | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
6Z6G
 
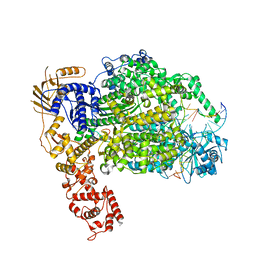 | | Cryo-EM structure of La Crosse virus polymerase at pre-initiation stage | | Descriptor: | 3'vRNA 1-16, 5'vRNA 1-10, 5'vRNA 9-16, ... | | Authors: | Arragain, B, Effantin, G, Gerlach, P, Reguera, J, Schoehn, G, Cusack, S, Malet, H. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
5NFZ
 
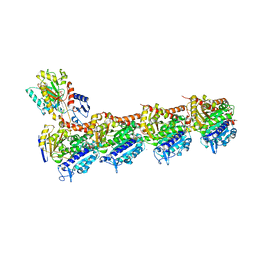 | | TUBULIN-MTC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-(2,3,4-trimethoxyphenyl)cyclohepta-2,4,6-trien-1-one, CALCIUM ION, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
5NGJ
 
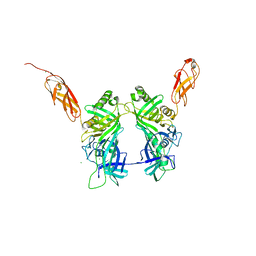 | | Crystal structure of pb6, major tail tube protein of bacteriophage T5 | | Descriptor: | CHLORIDE ION, Tail tube protein | | Authors: | Arnaud, C.-A, Effantin, G, Vives, C, Engilberge, S, Bacia, M, Boulanger, P, Girard, E, Schoehn, G, Breyton, C. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection.
Nat Commun, 8, 2017
|
|
5NG1
 
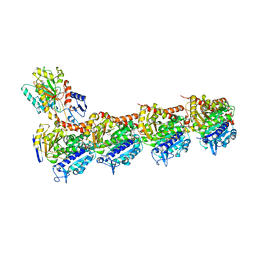 | | TUBULIN-MTC-zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, (2~{Z},4~{E})-~{N}-[(~{S})-oxidanyl-[(1~{S},2~{E},5~{S},11~{R},17~{S},19~{R})-3,11,19-trimethyl-7,13-bis(oxidanylidene)-6,21-dioxabicyclo[15.3.1]henicos-2-en-5-yl]methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
6HIN
 
 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, F conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIO
 
 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, I1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIS
 
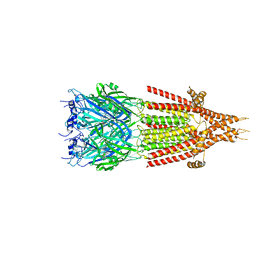 | | Mouse serotonin 5-HT3 receptor, tropisetron-bound, T conformation | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
6HIQ
 
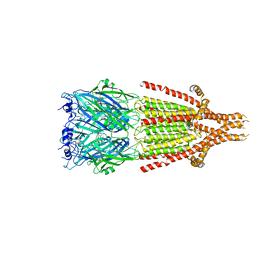 | | Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Polovinkin, L, Neumann, E, Schoehn, G, Nury, H. | | Deposit date: | 2018-08-30 | | Release date: | 2018-11-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions of the serotonin 5-HT3receptor.
Nature, 563, 2018
|
|
5MKC
 
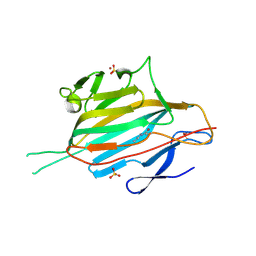 | | Crystal structure of the RrgA Jo.In complex | | Descriptor: | CALCIUM ION, Cell wall surface anchor family protein (Jo),Cell wall surface anchor family protein (In), NICKEL (II) ION, ... | | Authors: | Bonnet, J, Cartannaz, J, Tourcier, G, Contreras-Martel, C, Kleman, J.P, Fenel, D, Schoehn, G, Morlot, C, Vernet, T, Di Guilmi, A.M. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Autocatalytic association of proteins by covalent bond formation: a Bio Molecular Welding toolbox derived from a bacterial adhesin.
Sci Rep, 7, 2017
|
|
7OW8
 
 | | CryoEM structure of the ABC transporter BmrA E504A mutant in complex with ATP-Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
7PLM
 
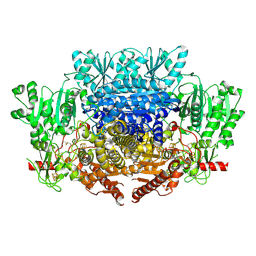 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
7ORJ
 
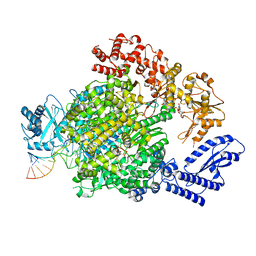 | | La Crosse virus polymerase at transcription capped RNA cleavage stage | | Descriptor: | 5' capped RNA, ADENOSINE-5'-TRIPHOSPHATE, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*CP*AP*AP*G)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORI
 
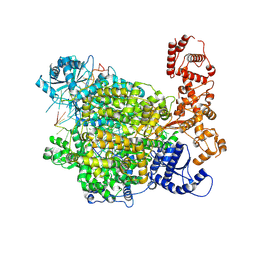 | | La Crosse virus polymerase at replication late-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORN
 
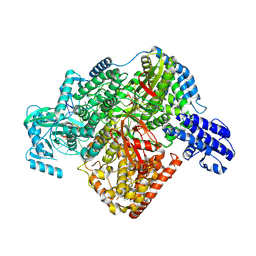 | | La Crosse virus polymerase at replication initiation stage | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, La Crosse virus polymerase, MAGNESIUM ION, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORL
 
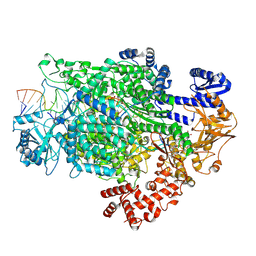 | | La Crosse virus polymerase at transcription initiation stage | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORM
 
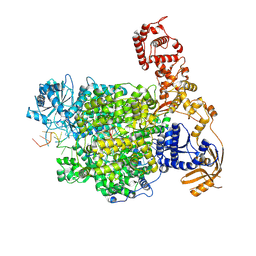 | | La Crosse virus polymerase at transcription early-elongation stage | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORK
 
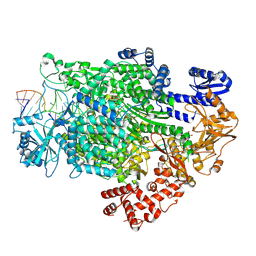 | | La Crosse virus polymerase in transcription mode with cleaved capped RNA entering the polymerase active site | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (5'-D(*(GTG))-R(P*A)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORO
 
 | | La Crosse virus polymerase at replication early-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7QOR
 
 | | Structure of beta-lactamase TEM-171 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Chojnowski, G, Lamzin, V, Egorov, A.M. | | Deposit date: | 2021-12-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6HX8
 
 | | Tubulin-STX3451 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dohle, W, Prota, A.E, Menchon, G, Hamel, E, Steinmetz, M.O, Potter, B.V.L. | | Deposit date: | 2018-10-16 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Tetrahydroisoquinoline Sulfamates as Potent Microtubule Disruptors: Synthesis, Antiproliferative and Antitubulin Activity of Dichlorobenzyl-Based Derivatives, and a Tubulin Cocrystal Structure.
ACS Omega, 4, 2019
|
|
6I2V
 
 | | Pilotin from Vibrio vulnificus type 2 secretion system, EpsS. | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Howard, S.P, Estrozi, L, Bertrand, Q, Contreras-Martel, C, Strozen, T, Job, V, Martins, A, Fenel, D, Schoehn, G, Dessen, A. | | Deposit date: | 2018-11-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and assembly of pilotin-dependent and -independent secretins of the type II secretion system.
Plos Pathog., 15, 2019
|
|
6I2N
 
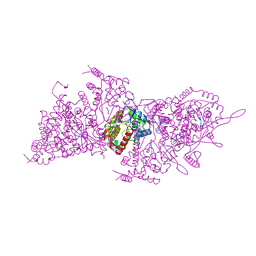 | | Helical RNA-bound Hantaan virus nucleocapsid | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*U)-3') | | Authors: | Arragain, B, Reguera, J, Desfosses, A, Gutsche, I, Schoehn, G, Malet, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms.
Elife, 8, 2019
|
|
8CK0
 
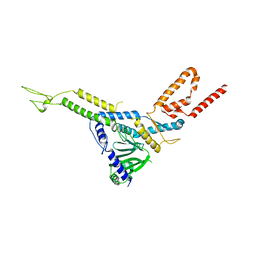 | |
