7B1C
 
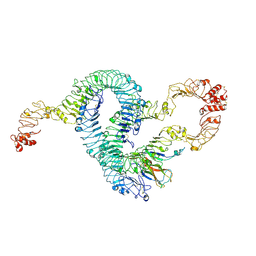 | | Cryo-EM of Aedes Aegypti Toll5A trimer bound to Spz1C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL013433-PA, ... | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
7B1B
 
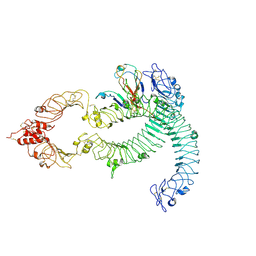 | | Cryo-EM of Aedes Aegypti Toll5A dimer bound to Spz1C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL013433-PA, ... | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
7B1D
 
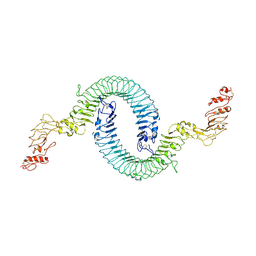 | | Cryo-EM of Aedes Aegypti Toll5A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
1PQF
 
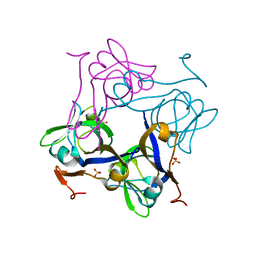 | | Glycine 24 to Serine mutation of aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYQ
 
 | | Unprocessed Aspartate Decarboxylase Mutant, with Alanine inserted at position 24 | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PPY
 
 | | Native precursor of pyruvoyl dependent Aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase precursor, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYU
 
 | | Processed Aspartate Decarboxylase Mutant with Ser25 mutated to Cys | | Descriptor: | Aspartate 1-decarboxylase alfa chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PQE
 
 | | S25A mutant of pyruvoyl dependent aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PT0
 
 | | Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase with an Alanine insertion at position 26 | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-22 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PT1
 
 | | Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase with Histidine 11 Mutated to Alanine | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-22 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PQH
 
 | | Serine 25 to Threonine mutation of aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, MALONIC ACID, SODIUM ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1GMN
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1GMO
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1GP9
 
 | | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Watanabe, K, Chirgadze, D.Y, Lietha, D, Gherardi, E, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping
J.Mol.Biol., 319, 2002
|
|
3KVU
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - T42S mutant in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KGV
 
 | |
3KVI
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42A mutant in complex with fluoro-acetate | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK, fluoroacetic acid | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KVZ
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thiesterase FlK - wild type FlK in complex with FAcCPan | | Descriptor: | (2R)-N-{3-[(5-fluoro-4-oxopentyl)amino]-3-oxopropyl}-2,4-dihydroxy-3,3-dimethylbutanamide, Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KW1
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - Wild type FlK in complex with FAcOPan | | Descriptor: | 2-({N-[(2S)-2,4-dihydroxy-3,3-dimethylbutanoyl]-beta-alanyl}amino)ethyl fluoroacetate, Fluoroacetyl-CoA thioesterase | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KV8
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - Wild type FlK in complex with fluoro-acetate | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK, fluoroacetic acid | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-29 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KX7
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - apo wild type FlK | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KUV
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42S mutant in complex with acetate. | | Descriptor: | ACETATE ION, Fluoroacetyl coenzyme A thioesterase | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
7PCH
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:2 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCQ
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:1 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
