5Z10
 
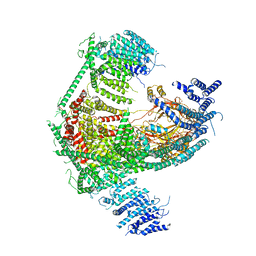 | | Structure of the mechanosensitive Piezo1 channel | | 分子名称: | Piezo-type mechanosensitive ion channel component 1 | | 著者 | Zhao, Q, Zhou, H, Chi, S, Wang, Y, Wang, J, Geng, J, Wu, K, Liu, W, Zhang, T, Dong, M.-Q, Wang, J, Li, X, Xiao, B. | | 登録日 | 2017-12-22 | | 公開日 | 2018-01-31 | | 最終更新日 | 2020-01-29 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | Structure and mechanogating mechanism of the Piezo1 channel.
Nature, 554, 2018
|
|
6R7G
 
 | | Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family | | 分子名称: | Coat protein, polyU RNA | | 著者 | Grinzato, A, Kandiah, E, Lico, C, Betti, C, Baschieri, S, Zanotti, G. | | 登録日 | 2019-03-28 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family.
Nat.Chem.Biol., 16, 2020
|
|
3KTL
 
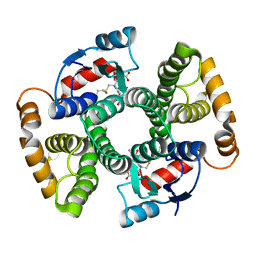 | | Crystal Structure of an I71A human GSTA1-1 mutant in complex with S-hexylglutathione | | 分子名称: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | 著者 | Achilonu, I.A, Gildenhuys, S, Fernandes, M.A, Fanucchi, S, Dirr, H.W. | | 登録日 | 2009-11-25 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The role of a topologically conserved isoleucine in glutathione transferase structure, stability and function.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2YPG
 
 | | Haemagglutinin of 1968 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Liu, J, Xiong, X, Haire, L.F, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | 登録日 | 2012-10-30 | | 公開日 | 2012-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8HAE
 
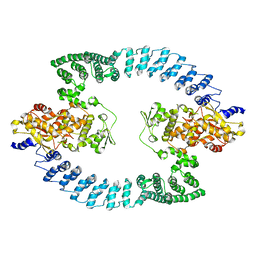 | | Cryo-EM structure of HACE1 dimer | | 分子名称: | E3 ubiquitin-protein ligase HACE1 | | 著者 | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J, Machida, S. | | 登録日 | 2022-10-26 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.55 Å) | | 主引用文献 | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
6XKR
 
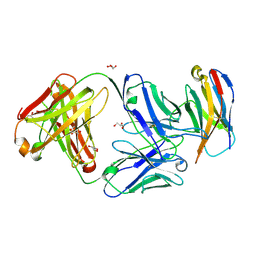 | | Structure of Sasanlimab Fab in complex with PD-1 | | 分子名称: | GLYCEROL, Programmed cell death protein 1, Sasanlimab Fab Heavy chain, ... | | 著者 | Kimberlin, C.R, Chin, S.M. | | 登録日 | 2020-06-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Pharmacologic Properties and Preclinical Activity of Sasanlimab, A High-affinity Engineered Anti-Human PD-1 Antibody.
Mol.Cancer Ther., 19, 2020
|
|
5KUF
 
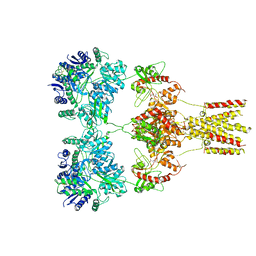 | | GluK2EM with 2S,4R-4-methylglutamate | | 分子名称: | 2S,4R-4-METHYLGLUTAMATE, Glutamate receptor ionotropic, kainate 2 | | 著者 | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | 登録日 | 2016-07-13 | | 公開日 | 2016-09-07 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
7M3V
 
 | |
5KUH
 
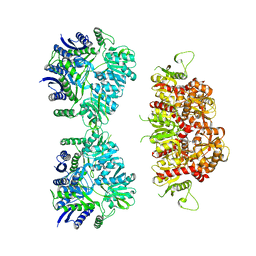 | | GluK2EM with LY466195 | | 分子名称: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | 著者 | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | 登録日 | 2016-07-13 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (11.6 Å) | | 主引用文献 | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
8H8X
 
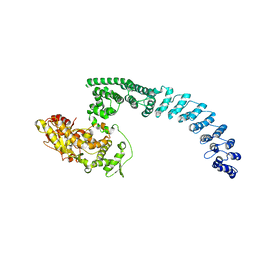 | | Cryo-EM structure of HACE1 monomer | | 分子名称: | E3 ubiquitin-protein ligase HACE1 | | 著者 | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | 登録日 | 2022-10-24 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8SJQ
 
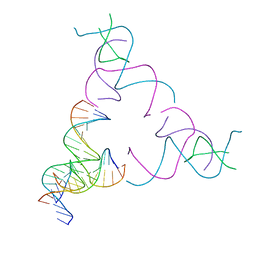 | | [3T16] Self-assembling right-handed tensegrity triangle with 16 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*GP*CP*CP*TP*GP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (6.19 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJR
 
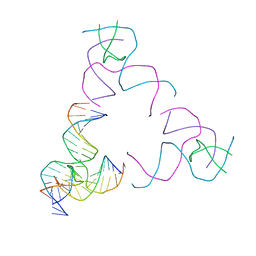 | | [3T17] Self-assembling right-handed tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (5.25 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJU
 
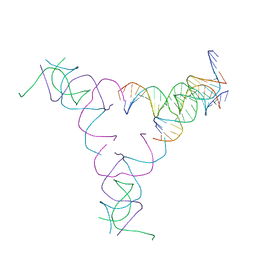 | | [4T17] Self-assembling right-handed four-turn tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (25-MER), DNA (5'-D(*GP*AP*AP*AP*AP*AP*CP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.14 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJM
 
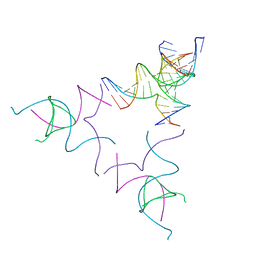 | | [3T12] Self-assembling left-handed tensegrity triangle with 12 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*AP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*TP*GP*CP*GP*AP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (8.08 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJO
 
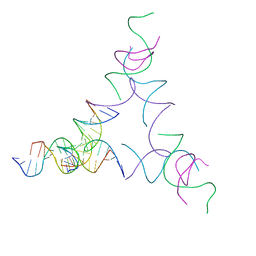 | | [3T14] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(P*CP*AP*CP*GP*TP*GP*GP*AP*CP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*AP*GP*CP*TP*CP*AP*GP*CP*CP*TP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*AP*GP*TP*CP*TP*CP*CP*AP*CP*GP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.08 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJN
 
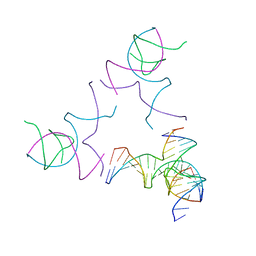 | | [3T13] Self-assembling left-handed tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*GP*AP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.3 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJS
 
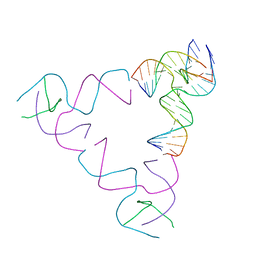 | | [3T18] Self-assembling right-handed tensegrity triangle with 18 interjunction base pairs and P63 symmetry | | 分子名称: | DNA (5'-D(*CP*AP*GP*AP*GP*CP*CP*TP*GP*AP*CP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (6.31 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJW
 
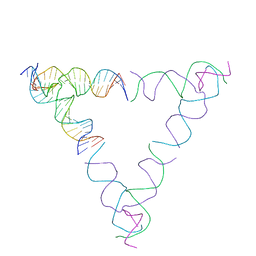 | | [4T28] Self-assembling right-handed tensegrity triangle with 28 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (28-MER), DNA (5'-D(*GP*AP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*TP*AP*CP*TP*GP*AP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*AP*GP*TP*GP*GP*CP*AP*GP*T)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (7.64 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJT
 
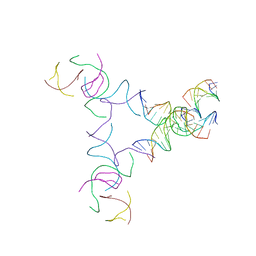 | | [3T14+10] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and a 10 bp linker with R3 symmetry | | 分子名称: | DNA (5'-D(*AP*CP*CP*TP*CP*CP*TP*GP*AP*GP*GP*TP*CP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*AP*CP*TP*CP*TP*GP*CP*TP*A)-3'), DNA (5'-D(*GP*TP*TP*AP*GP*CP*AP*GP*AP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (9.38 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJV
 
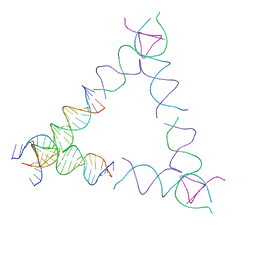 | | [4T24] Self-assembling left-handed tensegrity triangle with 24 interjunction base pairs and R3 symmetry | | 分子名称: | DNA (5'-D(P*CP*TP*TP*GP*TP*AP*GP*TP*CP*TP*CP*AP*CP*CP*AP*CP*TP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*CP*AP*CP*TP*CP*CP*TP*GP*AP*GP*AP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*CP*AP*TP*CP*AP*CP*AP*GP*TP*GP*GP*AP*CP*TP*AP*CP*AP*AP*G)-3'), ... | | 著者 | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | 登録日 | 2023-04-18 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (8.59 Å) | | 主引用文献 | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6LEO
 
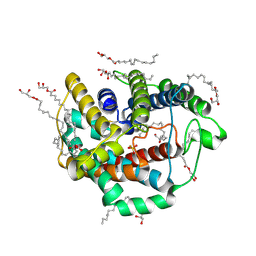 | | Crystal structure of thiosulfate transporter YeeE from Spirochaeta thermophila | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | 著者 | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Takeuchi, A, Uchino, S, Sugano, Y. | | 登録日 | 2019-11-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-30 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
6LEP
 
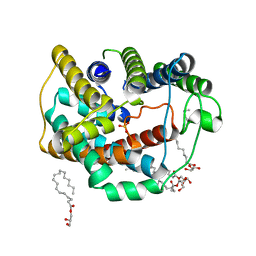 | | Crystal structure of thiosulfate transporter YeeE inactive mutant - C91A | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | 著者 | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Sugano, Y, Takeuchi, A, Uchino, S. | | 登録日 | 2019-11-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
4UFQ
 
 | | Structure of a novel Hyaluronidase (Hyal_Sk) from Streptomyces koganeiensis. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Gavira, J.A, Messina, L, Pernagallo, S, Unciti-Broceta, J.D, Conejero-Muriel, M, Diaz-Mochon, J.J, Vaccaro, S, Caruso, S, Musumeci, L, Bisicchia, S, Di Pasquale, R. | | 登録日 | 2015-03-18 | | 公開日 | 2016-04-13 | | 最終更新日 | 2017-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Identification and Characterization of a Bacterial Hyaluronidase and its Production in Recombinant Form.
FEBS Lett., 590, 2016
|
|
3RAO
 
 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987. | | 分子名称: | Putative Luciferase-like Monooxygenase, SULFATE ION | | 著者 | Domagalski, M.J, Chruszcz, M, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-03-28 | | 公開日 | 2011-05-11 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987.
To be Published
|
|
4UQ6
 
 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | 分子名称: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | 著者 | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
