7OSR
 
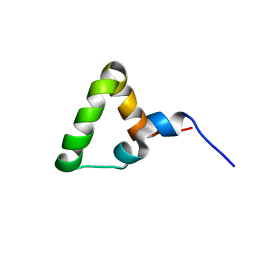 | |
7OSW
 
 | |
4UMG
 
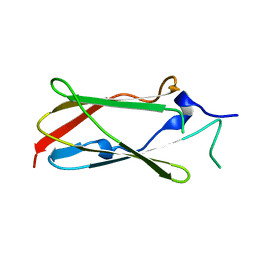 | | Crystal structure of the Lin-41 filamin domain | | Descriptor: | PROTEIN LIN-41 | | Authors: | Tocchini, C, Keusch, J.J, Miller, S.B, Finger, S, Gut, H, Stadler, M, Ciosk, R. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Trim-Nhl Protein Lin-41 Controls the Onset of Developmental Plasticity in Caenorhabditis Elegans.
Plos Genet., 10, 2014
|
|
1L3H
 
 | | NMR structure of P41icf, a potent inhibitor of human cathepsin L | | Descriptor: | MHC CLASS II-ASSOCIATED P41 INVARIANT CHAIN FRAGMENT (P41icf) | | Authors: | Chiva, C, Barthe, P, Codina, A, Giralt, E. | | Deposit date: | 2002-02-27 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Synthesis and NMR structure of P41ICF, a potent inhibitor of human cathepsin L
J.Am.Chem.Soc., 125, 2003
|
|
1YKR
 
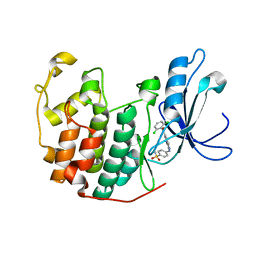 | | Crystal structure of cdk2 with an aminoimidazo pyridine inhibitor | | Descriptor: | 4-{[6-(2,6-DICHLOROBENZOYL)IMIDAZO[1,2-A]PYRIDIN-2-YL]AMINO}BENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Hamdouchi, C, Zhong, B, Mendoza, J, Jaramillo, C, Zhang, F, Brooks, H.B. | | Deposit date: | 2005-01-18 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a new class of highly selective aminoimidazo[1,2-a]pyridine-based inhibitors of cyclin dependent kinases
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7QCX
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, apo form | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
7QCY
 
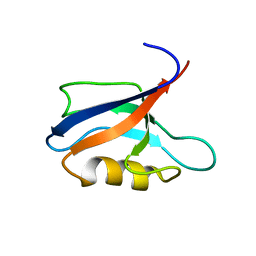 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, complexed with RA-GEF2 peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
4V5T
 
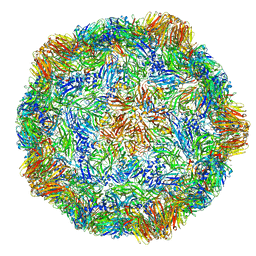 | | X-ray structure of the Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | Deposit date: | 2011-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
2JQC
 
 | | A L-amino acid mutant of a D-amino acid containing conopeptide | | Descriptor: | L-mr12 | | Authors: | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Purification and structural characterization of a d-amino acid-containing conopeptide, conomarphin, from Conus marmoreus
Febs J., 275, 2008
|
|
1G9I
 
 | | CRYSTAL STRUCTURE OF BETA-TRYSIN COMPLEX IN CYCLOHEXANE | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Zhu, G, Huang, Q, Zhu, Y, Li, Y, Chi, C, Tang, Y. | | Deposit date: | 2000-11-24 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray study on an artificial mung bean inhibitor complex with bovine beta-trypsin in neat cyclohexane.
Biochim.Biophys.Acta, 1546, 2001
|
|
1QUW
 
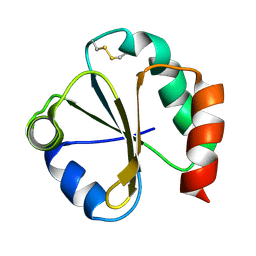 | | SOLUTION STRUCTURE OF THE THIOREDOXIN FROM BACILLUS ACIDOCALDARIUS | | Descriptor: | THIOREDOXIN | | Authors: | Nicastro, G, de Chiara, C, Pedone, E, Tato, M, Rossi, M. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a novel thioredoxin from Bacillus acidocaldarius possible determinants of protein stability.
Eur.J.Biochem., 267, 2000
|
|
3MYW
 
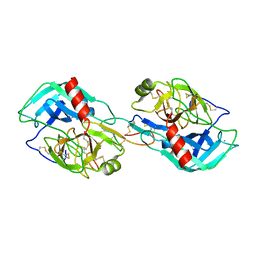 | | The Bowman-Birk type inhibitor from mung bean in ternary complex with porcine trypsin | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Engh, R.A, Bode, W, Huber, R, Lin, G, Chi, C. | | Deposit date: | 2010-05-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 0.25-nm X-ray structure of the Bowman-Birk-type inhibitor from mung bean in ternary complex with porcine trypsin.
Eur.J.Biochem., 212, 1993
|
|
1HNR
 
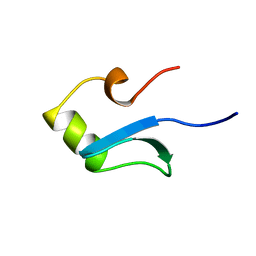 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1HNS
 
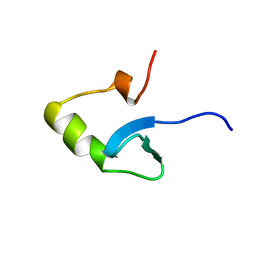 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
7C0N
 
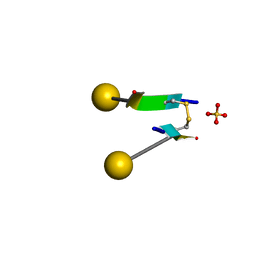 | | Crystal structure of a self-assembling galactosylated peptide homodimer | | Descriptor: | SULFATE ION, Self-assembling galactosylated tyrosine-rich peptide, beta-D-galactopyranose | | Authors: | He, C, Wu, S, Chi, C, Zhang, W, Ma, M, Lai, L, Dong, S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Glycopeptide Self-Assembly Modulated by Glycan Stereochemistry through Glycan-Aromatic Interactions.
J.Am.Chem.Soc., 142, 2020
|
|
2B1U
 
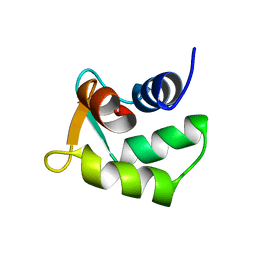 | | Solution structure of Calmodulin-like Skin Protein C terminal domain | | Descriptor: | Calmodulin-like protein 5 | | Authors: | Babini, E, Bertini, I, Capozzi, F, Chirivino, E, Luchinat, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-16 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural and Dynamic Characterization of the EF-Hand Protein CLSP.
Structure, 14, 2006
|
|
1SBW
 
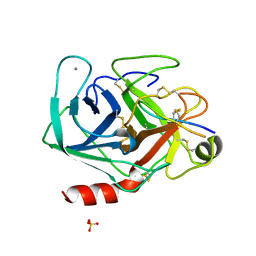 | | CRYSTAL STRUCTURE OF MUNG BEAN INHIBITOR LYSINE ACTIVE FRAGMENT COMPLEX WITH BOVINE BETA-TRYPSIN AT 1.8A RESOLUTION | | Descriptor: | CALCIUM ION, PROTEIN (BETA-TRYPSIN), PROTEIN (MUNG BEAN INHIBITOR LYSIN ACTIVE FRAGMENT), ... | | Authors: | Huang, Q, Zhu, Y, Chi, C, Tang, Y. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mung bean inhibitor lysine active fragment complex with bovine beta-trypsin at 1.8A resolution.
J.Biomol.Struct.Dyn., 16, 1999
|
|
2JQB
 
 | | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5 | | Descriptor: | Conomarphin | | Authors: | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | Deposit date: | 2007-05-31 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5
To be Published
|
|
2AJM
 
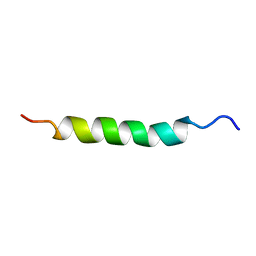 | | NMR structure of the in-plane membrane anchor domain [1-28] of the monotopic NonStructural Protein 5A (NS5A) from the Bovine Viral Diarrhea Virus (BVDV) | | Descriptor: | Nonstructural protein 5A | | Authors: | Sapay, N, Montserret, R, Chipot, C, Brass, V, Moradpour, D, Deleage, G, Penin, F. | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus.
Biochemistry, 45, 2006
|
|
6ES4
 
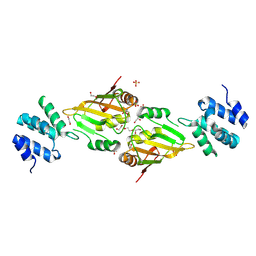 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
8XZ2
 
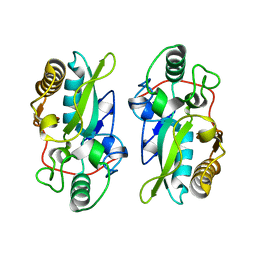 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
3ZYP
 
 | | Cellulose induced protein, Cip1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CIP1, ... | | Authors: | Jacobson, F, Karkehabadi, S, Hansson, H, Goedegebuur, F, Wallace, L, Mitchinson, C, Piens, K, Stals, I, Sandgren, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Core Domain of a Cellulose Induced Protein (Cip1) from Hypocrea Jecorina, at 1.5 A Resolution.
Plos One, 8, 2013
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6EOK
 
 | | Crystal structure of E. coli L-asparaginase II | | Descriptor: | L-asparaginase 2, ZINC ION | | Authors: | Cerofolini, L, Giuntini, S, Carlon, A, Ravera, E, Calderone, V, Fragai, M, Parigi, G, Luchinat, C. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of PEGylated Asparaginase: New Opportunities from NMR Analysis of Large PEGylated Therapeutics.
Chemistry, 25, 2019
|
|
6LLK
 
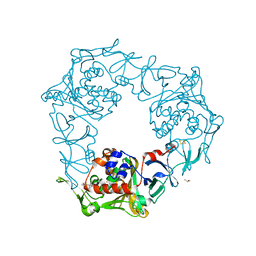 | | Biphenyl-2,2',3-triol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-hydroxyphenyl)benzene-1,2-diol, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-23 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biphenyl-2,2',3-triol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase
To Be Published
|
|
