2A5K
 
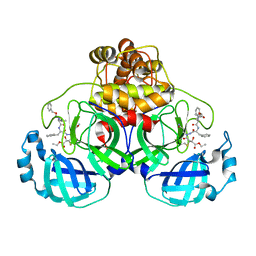 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5A
 
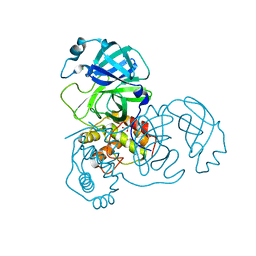 | | Crystal structure of unbound SARS coronavirus main peptidase in the space group C2 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like peptidase, CHLORIDE ION | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
2A15
 
 | | X-ray Crystal Structure of RV0760 from Mycobacterium Tuberculosis at 1.68 Angstrom Resolution | | Descriptor: | HYPOTHETICAL PROTEIN Rv0760c, NICOTINAMIDE | | Authors: | Garen, C.R, Cherney, M.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase
Biochim.Biophys.Acta, 1784, 2008
|
|
2ASF
 
 | | Crystal structure of the conserved hypothetical protein Rv2074 from Mycobacterium tuberculosis 1.6 A | | Descriptor: | CITRIC ACID, Hypothetical protein Rv2074, SODIUM ION | | Authors: | Biswal, B.K, Au, K, Cherney, M.M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular structure of Rv2074, a probable pyridoxine 5'-phosphate oxidase from Mycobacterium tuberculosis, at 1.6 angstroms resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2BCD
 
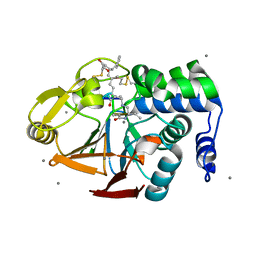 | | X-ray crystal structure of Protein Phosphatase-1 with the marine toxin motuporin bound | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, MOTUPORIN, ... | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-19 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
2BDX
 
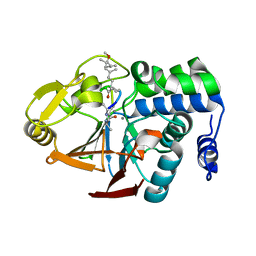 | | X-ray Crystal Structure of dihydromicrocystin-LA bound to Protein Phosphatase-1 | | Descriptor: | DIHYDROMICROCYSTIN-LA, MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-gamma catalytic subunit | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-21 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
2GT7
 
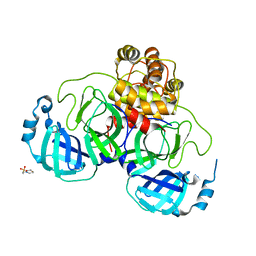 | | Crystal structure of SARS coronavirus main peptidase at pH 6.0 in the space group P21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2GT8
 
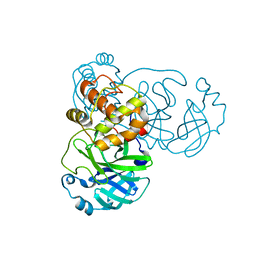 | | Crystal structure of SARS coronavirus main peptidase (with an additional Ala at the N-terminus of each protomer) in the space group P43212 | | Descriptor: | 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2GK1
 
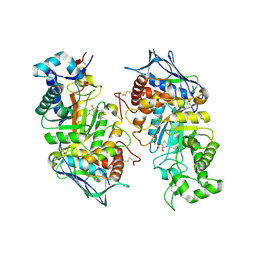 | | X-ray crystal structure of NGT-bound HexA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase subunit alpha, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystallographic Structure of Human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of G(M2) Ganglioside Hydrolysis.
J.Mol.Biol., 359, 2006
|
|
2GTB
 
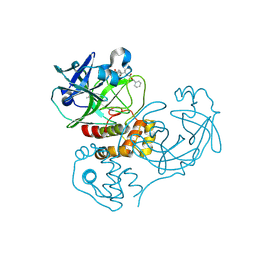 | | Crystal structure of SARS coronavirus main peptidase (with an additional Ala at the N-terminus of each protomer) inhibited by an aza-peptide epoxide in the space group P43212 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like proteinase, ACETIC ACID | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2GKE
 
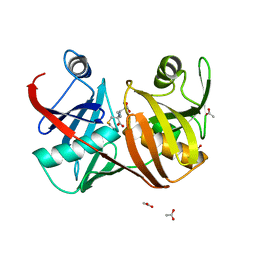 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase, ... | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-01 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GJX
 
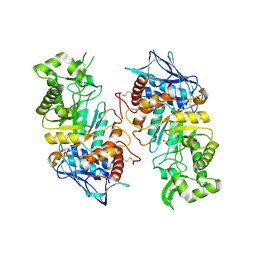 | | Crystallographic structure of human beta-Hexosaminidase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase alpha chain, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic structure of human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss
of GM2 Ganglioside Hydrolysis
J.Mol.Biol., 359, 2006
|
|
2D41
 
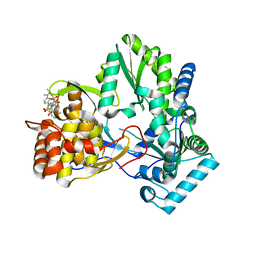 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | Descriptor: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-05 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3Z
 
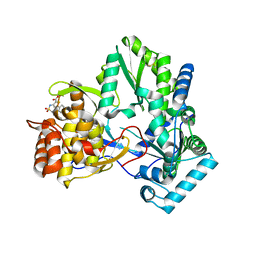 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2CXV
 
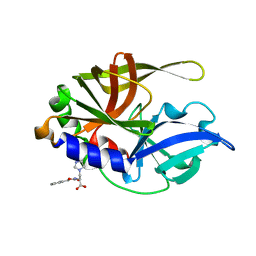 | | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-Derived betaLactone: Selective Crystallization and High-resolution Structure of the His-102 Adduct | | Descriptor: | N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Probable protein P3C | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
2GKJ
 
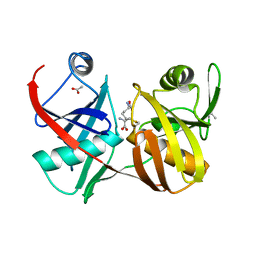 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor DL-AZIDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-02 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2D3U
 
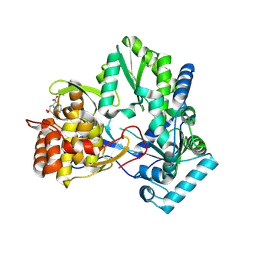 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2DJ5
 
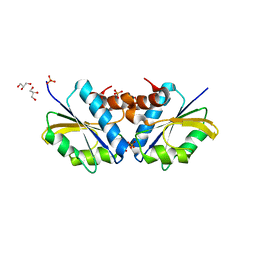 | | Crystal Structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Cherney, M.M, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Vitamin B(12) Biosynthetic Cobaltochelatase, CbiX (S), from Archaeoglobus Fulgidus
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
2HRV
 
 | | 2A CYSTEINE PROTEINASE FROM HUMAN RHINOVIRUS 2 | | Descriptor: | 2A CYSTEINE PROTEINASE, ZINC ION | | Authors: | Petersen, J.F.W, Cherney, M.M, Liebig, H.-D, Skern, T, Kuechler, E, James, M.N.G. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis.
EMBO J., 18, 1999
|
|
2ATO
 
 | | Crystal structure of Human Cathepsin K in complex with myocrisin | | Descriptor: | (S)-(1,2-DICARBOXYETHYLTHIO)GOLD, Cathepsin K, SULFATE ION | | Authors: | Weidauer, E, Yasuda, Y, Biswal, B.K, Kerr, L.D, Cherney, M.M, Gordon, R.E, James, M.N.G, Bromme, D. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of disease-modifying anti-rheumatic drugs (DMARDs) on the activities of rheumatoid arthritis-associated cathepsins K and S.
Biol.Chem., 388, 2007
|
|
