7LNI
 
 | | SeMet CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | 著者 | Horton, J.R, Cheng, X, Zhou, J. | | 登録日 | 2021-02-07 | | 公開日 | 2021-05-19 | | 最終更新日 | 2021-07-14 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LT5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Cofactor SAH | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | 著者 | Horton, J.R, Cheng, X, Zhou, J. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LNJ
 
 | |
3KV5
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV9
 
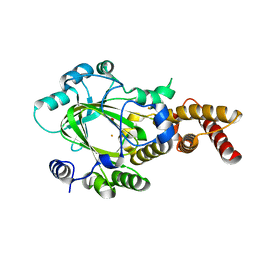 | | Structure of KIAA1718 Jumonji domain | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8SST
 
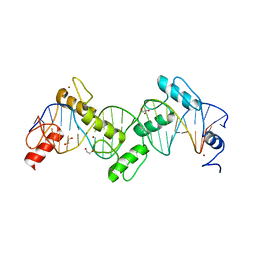 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) K365T Mutant Complexed with 23mer | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSU
 
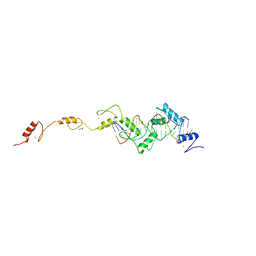 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSQ
 
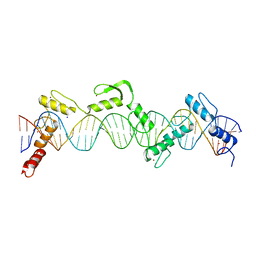 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | 分子名称: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | 分子名称: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSS
 
 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
3KVB
 
 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | 分子名称: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV4
 
 | | Structure of PHF8 in complex with histone H3 | | 分子名称: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4F7Z
 
 | | Conformational dynamics of exchange protein directly activated by cAMP | | 分子名称: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | 著者 | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | 登録日 | 2012-05-16 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
2G45
 
 | | Co-crystal structure of znf ubp domain from the deubiquitinating enzyme isopeptidase T (isot) in complex with ubiquitin | | 分子名称: | CHLORIDE ION, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 5, ... | | 著者 | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | 登録日 | 2006-02-21 | | 公開日 | 2006-04-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2G43
 
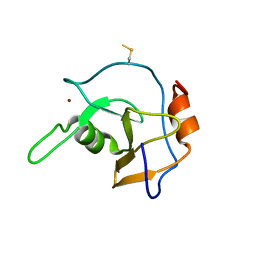 | | Structure of the ZNF UBP domain from deubiquitinating enzyme isopeptidase T (IsoT) | | 分子名称: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ZINC ION | | 著者 | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | 登録日 | 2006-02-21 | | 公開日 | 2006-04-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2H11
 
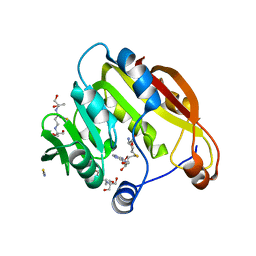 | | Amino-terminal Truncated Thiopurine S-Methyltransferase Complexed with S-Adenosyl-L-Homocysteine | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOCYANATE ION, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2006-05-15 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis of allele variation of human thiopurine-S-methyltransferase.
Proteins, 67, 2007
|
|
2M2D
 
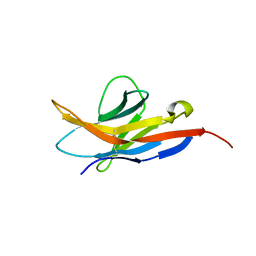 | | Human programmed cell death 1 receptor | | 分子名称: | Programmed cell death protein 1 | | 著者 | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | 登録日 | 2012-12-18 | | 公開日 | 2013-02-27 | | 最終更新日 | 2013-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
2MB5
 
 | |
3MO0
 
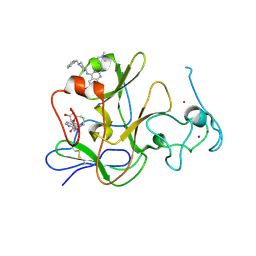 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E11 | | 分子名称: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | 著者 | Chang, Y, Horton, J.R, Cheng, X. | | 登録日 | 2010-04-22 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
3MO5
 
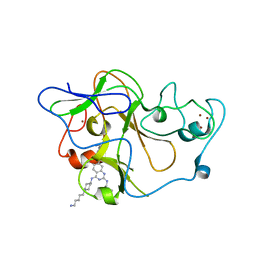 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E72 | | 分子名称: | 7-[(5-aminopentyl)oxy]-N~4~-[1-(5-aminopentyl)piperidin-4-yl]-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | 著者 | Chang, Y, Horton, J.R, Cheng, X. | | 登録日 | 2010-04-22 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
3MO2
 
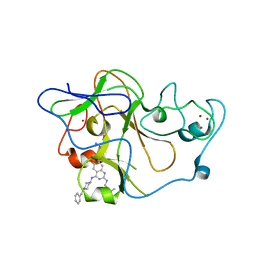 | | human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E67 | | 分子名称: | 7-[(5-aminopentyl)oxy]-N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | 著者 | Chang, Y, Horton, J.R, Cheng, X. | | 登録日 | 2010-04-22 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
4LT5
 
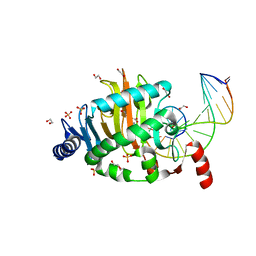 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | 著者 | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | 登録日 | 2013-07-23 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.893 Å) | | 主引用文献 | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
4M9E
 
 | | Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA | | 分子名称: | ACETATE ION, DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), ... | | 著者 | Liu, Y, Olanrewaju, Y.O, Blumenthal, R.M, Zhang, X, Cheng, X. | | 登録日 | 2013-08-14 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Structural basis for Klf4 recognition of methylated DNA.
Nucleic Acids Res., 42, 2014
|
|
