2AOI
 
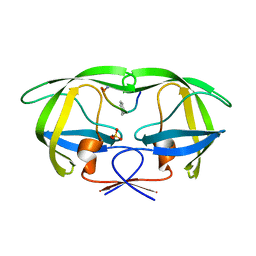 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | Descriptor: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOE
 
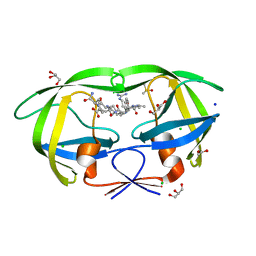 | | crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog CA-P2 | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOF
 
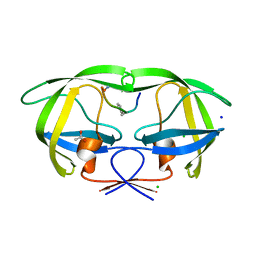 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P1-P6 | | Descriptor: | ACETIC ACID, CHLORIDE ION, PEPTIDE INHIBITOR, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOH
 
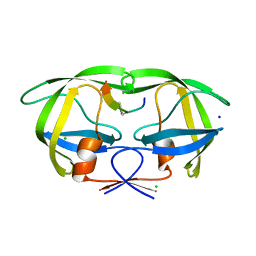 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | Descriptor: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
4D8P
 
 | | Structural and functional studies of the trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) molecule | | Descriptor: | GLYCEROL, HLA-DQA1 protein, Peptide from Gamma-gliadin,HLA class II histocompatibility antigen, ... | | Authors: | Kim, C.-Y, Hotta, K, Mathews, I.I, Chen, X. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and functional studies of trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) protein molecule
J.Biol.Chem., 287, 2012
|
|
8CIE
 
 | | Crystal structure of the human CDKL5 kinase domain with compound YL-354 | | Descriptor: | 4-[[3,5-bis(fluoranyl)phenyl]carbonylamino]-~{N}-piperidin-4-yl-1~{H}-pyrazole-3-carboxamide, Cyclin-dependent kinase-like 5, SULFATE ION | | Authors: | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.
Acs Chem Neurosci, 14, 2023
|
|
1WM8
 
 | | Solution Structure of BmP03 from the Venom of Scorpion Buthus martensii Karsch, 10 structures | | Descriptor: | Neurotoxin BmP03 | | Authors: | Wu, H, He, F, Li, Y, Wu, G, Cao, C, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of BmP03 from Venom of Scorpion Buthus martensii Karsch
Acta Chim.Sinica, 58, 2000
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
1WWN
 
 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1MPW
 
 | | Molecular Recognition in (+)-a-Pinene Oxidation by Cytochrome P450cam | | Descriptor: | (+)-alpha-Pinene, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Bell, S.G, Chen, X, Sowden, R.J, Xu, F, Willams, J.N, Wong, L.-L, Rao, Z. | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Molecular recognition in (+)-alpha-pinene oxidation by cytochrome P450cam
J.Am.Chem.Soc., 125, 2003
|
|
8KH6
 
 | | Crystal structure of FGFR4 kinase domain with 8r | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[5-bromanyl-6-(hydroxymethyl)-3-methoxy-pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH9
 
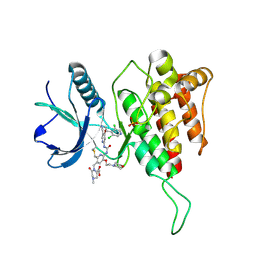 | | Crystal structure of FGFR4(V550M) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH7
 
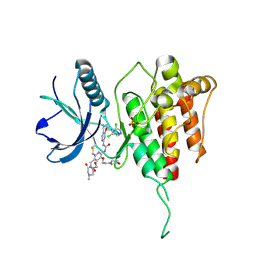 | | Crystal structure of FGFR4 kinase domain with 8zc | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
1N25
 
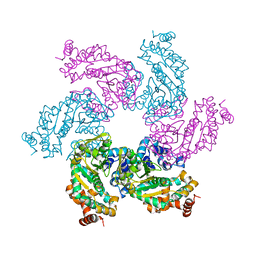 | | Crystal structure of the SV40 Large T antigen helicase domain | | Descriptor: | Large T Antigen, ZINC ION | | Authors: | Li, D, Zhao, R, Lilyestrom, W, Gai, D, Zhang, R, DeCaprio, J.A, Fanning, E, Jochimiak, A, Szakonyi, G, Chen, X.S. | | Deposit date: | 2002-10-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen
Nature, 423, 2003
|
|
8GS5
 
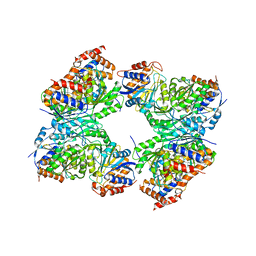 | | Crystal structure of a constitutively active mutant of human IDH3 holoenzyme in apo form | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.486 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRG
 
 | |
8GRH
 
 | | Crystal structure of a constitutively active mutant of the alpha gamma heterodimer of human IDH3 in complex with CIT | | Descriptor: | CITRIC ACID, Human IDH3 alpha subunit, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRU
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ICT, NAD and Ca | | Descriptor: | CALCIUM ION, Human IDH3 alpha subunit, ISOCITRIC ACID, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRB
 
 | |
1R22
 
 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB (C14S/C61S/C121S mutant) in the Zn2alpha5-form | | Descriptor: | Transcriptional repressor smtB, ZINC ION | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R1U
 
 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the apo-form | | Descriptor: | repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R23
 
 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the Zn1-form (one Zn(II) per dimer) | | Descriptor: | Transcriptional repressor smtB, ZINC ION | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
8KH8
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
1R1T
 
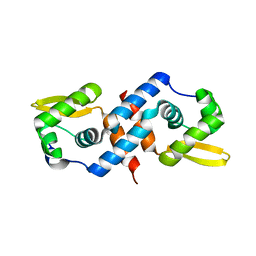 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the apo-form | | Descriptor: | Transcriptional repressor smtB | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
4GQH
 
 | | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies | | Descriptor: | Capsid protein | | Authors: | Li, X.Y, Song, B.A, Hu, D.Y, Chen, X, Wang, Z.C, Zeng, M.J, Yu, D.D, Chen, Z, Jin, L.H, Yang, S. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies
To be Published
|
|
