3ZPA
 
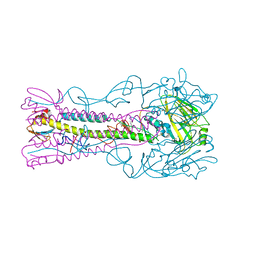 | | INFLUENZA VIRUS (VN1194) H5 I155F mutant HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ | | Authors: | Liu, J, Chen, Z, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-27 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3MVT
 
 | | Crystal structure of apo mADA at 2.2A resolution | | Descriptor: | Adenosine deaminase, CHLORIDE ION, GLYCEROL | | Authors: | Niu, W, Shu, Q, Chen, Z, Mathews, S, Di Cera, E, Frieden, C. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The role of Zn2+ on the structure and stability of murine adenosine deaminase.
J.Phys.Chem.B, 114, 2010
|
|
2M01
 
 | | Solution structure of Kunitz-type neurotoxin LmKKT-1a from scorpion venom | | Descriptor: | Protease inhibitor LmKTT-1a | | Authors: | Luo, F, Jiang, L, Liu, M, Chen, Z, Wu, Y. | | Deposit date: | 2012-10-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins.
Plos One, 8, 2013
|
|
4AKO
 
 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: E498L mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Silva, C.S, Chen, Z, Durao, P, Pereira, M.M, Todorovic, S, Hildebrandt, P, Martins, L.O, Lindley, P.F, Bento, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of Glu498 in the Dioxygen Reactivity of Cota-Laccase from Bacillus Subtilis.
Dalton Trans, 39, 2010
|
|
6JDS
 
 | |
6JDU
 
 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
5X0T
 
 | | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody 6H8 | | Descriptor: | 6H8 Fab fragment heavy chain, 6H8 Fab fragment light chain, Basigin | | Authors: | Lin, P, Zhang, M.-Y, Ye, S, Chen, X, Yu, X.-L, Zhang, R.-G, Zhu, P, Chen, Z.-N. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody
To Be Published
|
|
5X4G
 
 | | Crystal structure of Fab fragment of anti-CD147 monoclonal antibody 6H8 | | Descriptor: | 6H8 Fab heavy chain, 6H8 Fab light chain | | Authors: | Lin, P, Zhang, M.-Y, Chen, X, Ye, S, Yu, X.-L, Zhang, R.-G, Zhu, P, Chen, Z.-N. | | Deposit date: | 2017-02-13 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody
To Be Published
|
|
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2010-09-04 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
4ZVV
 
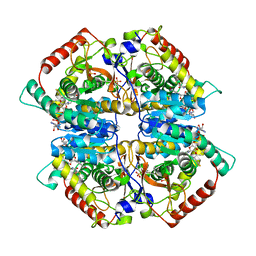 | | Lactate dehydrogenase A in complex with a trisubstituted piperidine-2,4-dione inhibitor GNE-140 | | Descriptor: | (2~{R})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Li, Y, Chen, Z, Eigenbrot, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition.
Nat.Chem.Biol., 12, 2016
|
|
4HZH
 
 | | Structure of recombinant Gla-domainless prothrombin mutant S525A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pozzi, N, Niu, W, Gohara, D.W, Chen, Z, Di Cera, E. | | Deposit date: | 2012-11-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of prothrombin reveals conformational flexibility and mechanism of activation.
J.Biol.Chem., 288, 2013
|
|
4DFI
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
4DFH
 
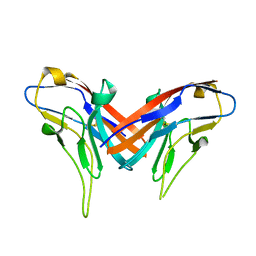 | | Crystal structure of cell adhesion molecule nectin-2/CD112 variable domain | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
7FDV
 
 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | Authors: | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | Deposit date: | 2021-07-18 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
5CI6
 
 | |
7XV6
 
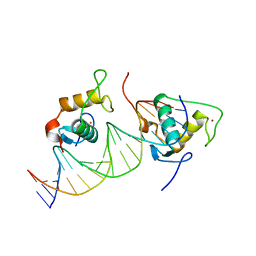 | |
7XV9
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain | | Descriptor: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV8
 
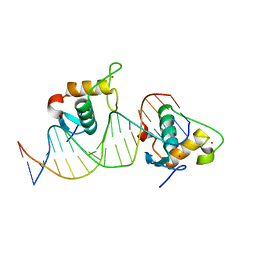 | | Crystal structure of the Human TR4 DNA-Binding Domain Homodimer Bound to DR1 Response Element | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), Nuclear receptor subfamily 2 group C member 2, ... | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
4FSJ
 
 | | Crystal structure of the virus like particle of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4GEE
 
 | | Pyrrolopyrimidine inhibitors of DNA gyrase B and topoisomerase IV, part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-yloxy)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Chen, Z, Tari, L.W. | | Deposit date: | 2012-08-01 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4FTS
 
 | | Crystal structure of the N363T mutant of the Flock House virus capsid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4FWE
 
 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
3MVI
 
 | | Crystal structure of holo mADA at 1.6 A resolution | | Descriptor: | Adenosine deaminase, GLYCEROL, ZINC ION | | Authors: | Niu, W, Shu, Q, Chen, Z, Mathews, S, Di Cera, E, Frieden, C. | | Deposit date: | 2010-05-04 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of Zn2+ on the structure and stability of murine adenosine deaminase.
J.Phys.Chem.B, 114, 2010
|
|
3CX8
 
 | |
6K15
 
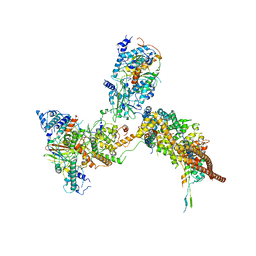 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
